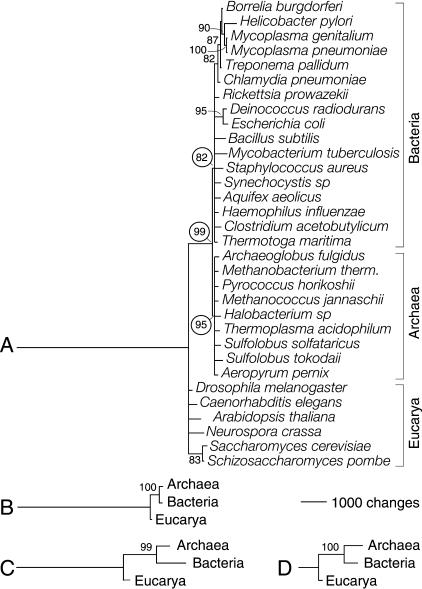
Figure 2.
Phylogenetic reconstruction of a universal tree. Phylogenetic relationships were inferred from genomic abundance values of SCOP 1.59 fold categories. Bootstrap support (BS) values >80% are shown above nodes. (A) Reduced phylogenetic tree reconstructed from fold occurrence (G) data. A total of 507 informative out of 536 total characters with 20 character states each were analyzed. Two most-parsimonious trees of 16,157 steps (CI = 0.625, RI = 0.486; g1 = -0.659; PTP test, P = 0.001) were retained after a heuristic search with tree-bisection-reconnection (TBR) branch swapping and 50 replicates of random addition sequence. The tree shown is congruent with the 50% majority-rule consensus. The null hypothesis of congruence could not be rejected when folds in the six structural classes were tested for homogeneity of data partitions (P = 0.498). (B) Tree reconstructed from fold occurrence data averaged across genomes in each organismal domain (Ḡ). Characters had 20 states, and 300 informative characters were analyzed. A single tree of 5885 steps (CI = 0.970, RI = 0.660; g1 = -0.702; PTP, P = 0.001) was retained after an exhaustive search. (C) Tree reconstructed from the fraction of genomes in each organismal domain that share individual folds (f). Characters had 17 states; 447 informative out of 507 total characters were analyzed. A single tree of 7603 steps (CI = 0.852, RI = 0.543; g1 = -0.559; PTP, P = 0.001) was retained after an exhaustive search. (D) Tree reconstructed as in C but from the subset of folds that is shared by the three organismal domains. Characters had 17 states, and 149 informative out of 246 total characters were analyzed. A single tree of 1601 steps (CI = 0.895, RI = 0.752; g1 = -0.672; PTP, P = 0.001) was retained after an exhaustive search.