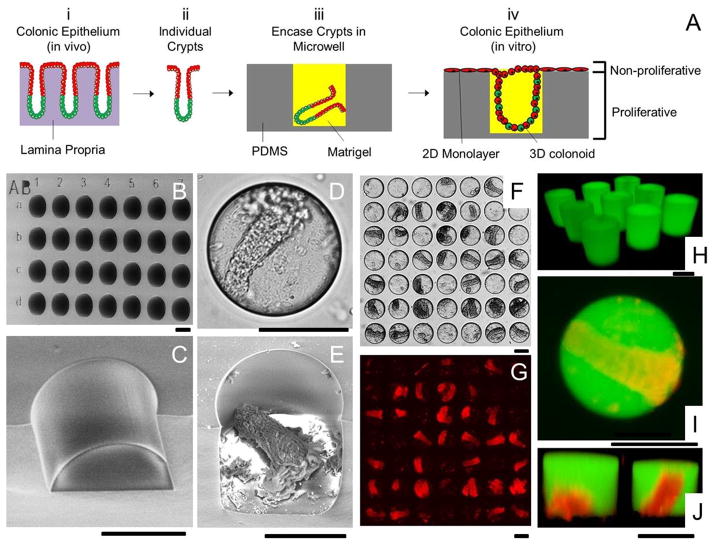
Figure 3.
Microwell arrays for combined 2D/3D culture of crypts. (A) Schematic of the 2D/3D culture strategy. (i) The colonic epithelium in vivo is composed of crypt subunits each in contact with a lamina propria. (ii) Individual crypts are isolated from a colon. (iii) Crypts are loaded into PDMS microwells and encased within Matrigel micropockets. (iv) In vitro culture of crypts in microwells generates colonic epithelium resembling the polarized architecture of crypts in vivo. (B) SEM image of the PDMS microwell array. The wells were labeled with shallow alphanumeric characters. (C) A close-up SEM image of a disrupted section of a microwell. (D–E) Brightfield (D) and SEM image (E) of a microwell with a loaded crypt. (F) Brightfield image showing 28 out of 49 wells loaded with crypts. (G) DsRed fluorescence image of F. (H) Fluorescence confocal image of Matrigel pockets formed in the microwell array. (I–J) Shown is a standard fluorescence image (I) and a confocal reconstructed Z slice (J) of crypts encapsulated within isolated Matrigel pockets in the microwell array. The panels display overlaid red/green fluorescence images. The Matrigel was mixed with 100 μg/mL fluorescein-dextran in images H through I. Scale bars = 100 μm for images in B–J.