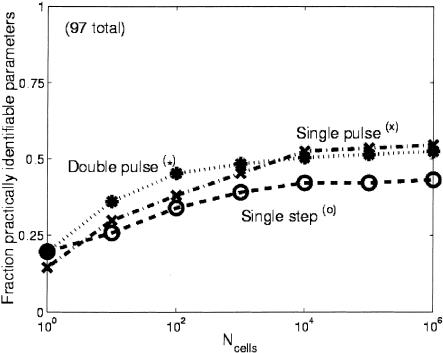
Figure 4.
Fraction of practically identifiable parameters (see Methods section for definition) vs. the number of cells sampled per time point (Ncells) for the three input perturbations. When only a few cells are sampled per time point, the effect of stochastic gene expression is the greatest, and each perturbation allows about the same number of parameters to be identified with statistical confidence. As more cells are sampled per time point, the noise is reduced and the double-pulse perturbation performs the best. When a very large number of cells are sampled, greatly reducing the impact of stochastic gene expression, the single pulse performs as well as or slightly better than the double pulse. The slight advantage of the single pulse in this limit is due to the slightly greater number of parameters that are a priori identifiable with that perturbation (Table 1).