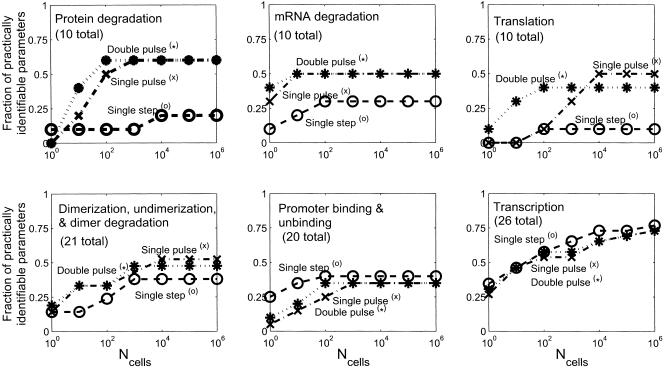
Figure 5.
Fraction of practically identifiable parameters (see Methods for definition) by biological function vs. the number of cells sampled per time point (Ncells) for the three input perturbations: For protein degradation and translation parameters, the different perturbations perform about equally well when Ncells is small (high noise), with the double pulse being superior at intermediate Ncells, and the double and single pulses performing about equally well at very high Ncells. For mRNA degradation, dimerization, undimerization, dimer degradation, promoter binding, and unbinding, both pulse perturbations perform about equally well. Interestingly, the step perturbation is more effective for identifying promoter binding and unbinding parameters. All three perturbations perform about equally well for the transcriptional parameters. The differing effectiveness for the perturbations for the different classes of parameters may result from their tendency to excite different time scales in the system.