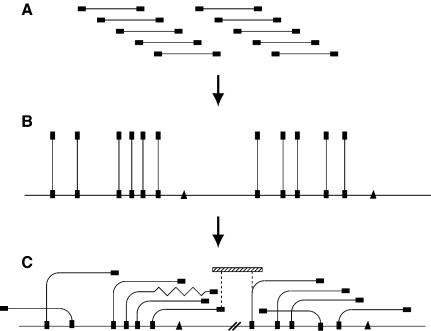
Figure 1.
Construction of HAPPily-anchored physical map. (A) Members of a genomic PAC library are end-sequenced (filled rectangles). (B) One end-sequence of each clone is HAPPY-mapped to establish its position in the genome; additional sequence-tagged sites (triangles) are also mapped. (C) Overlaps between nearby clones are determined, by using PCR to test each clone for its content of nearby mapped markers, creating contigs. Chimeric clones or deletions (zigzag line) become apparent. The orientations of contigs which are separated by uncloned portions (heavy parallel diagonal lines) are known from the HAPPY map, and additional linking clones (hatched rectangle) can be sought by screening the same or other libraries with the markers adjacent to the gap (dashed vertical lines).