Abstract
Nephronophthisis (NPHP), an autosomal recessive cystic kidney disease, is the most frequent genetic cause for end-stage renal failure in the first 3 decades of life. Mutations in 13 genes (NPHP1-NPHP11, AHI1, and CC2D2A) cause NPHP with ubiquitous expression of the corresponding proteins consistent with the multiorgan involvement of NPHP-related diseases. The genotype-phenotype correlation in these ciliopathies can be explained by gene locus heterogeneity, allelism, and the impact of modifier genes. In some NPHP-related ciliopathies, the nature of the recessive mutations determines disease severity. In order to define the genotypephenotype correlation more clearly, we evaluated a worldwide cohort of 440 patients from 365 families with NPHP-related ciliopathies, in whom both disease-causing alleles were identified. The phenotypes were ranked in the order of severity from degenerative to degenerative/ dysplastic to dysplastic. A genotype of 2 null alleles caused a range of phenotypes with an increasing order of severity of NPHP1, NPHP3, NPHP4, NPHP5, NPHP2, NPHP10, NPHP6 to AHI1. Only NPHP6 showed allelic influences on the phenotypes; the presence of 2 null mutations caused dysplastic phenotypes, whereas at least one missense allele rescued it to a milder degenerative phenotype. We also found 9 novel mutations in the NPHP genes. Thus, our studies have important implications for genetic counseling and planning of renal replacement therapy.
Keywords: cystic kidney, end-stage renal disease, genetic renal disease, human genetics, pediatric nephrology
Nephronophthisis (NPHP) is an autosomal recessive cystic kidney disease that represents the most frequent genetic cause of end-stage renal disease (ESRD) in the first 3 decades of life.1 So far, 13 genes (NPHP1–NPHP11, AHI1, and CC2D2A) have been identified that cause NPHP when mutated.2–15 Interestingly, NPHP can occur with isolated kidney involvement or in combination with diverse extrarenal manifestations. Specifically, it can be associated with: (i) retinal degeneration in Senior–Løken syndrome (SLSN); (ii) cerebellar vermis aplasia (CVA)/hypoplasia (CVH), retinal degeneration, and mental retardation in Joubert syndrome (JBTS); and (iii) dysplastic phenotypes of multiple organs leading to lethal Meckel–Gruber syndrome (MKS). The genotype–phenotype correlation in NPHP-related ciliopathies (NPHP-RC) seems to be governed by three genetic mechanisms: gene locus heterogeneity, allelism, and modifier genes. Although the ubiquitous expression of NPHP proteins corresponds to the multiorgan involvement in NPHP-RC, the pleiotropy of NPHP genes raises the importance of allelism as the nature of the mutations can also determine disease severity. Sometimes the presence of two null (protein-truncating/nonsense) mutations causes severe, early-onset, dysplastic, multiorgan phenotypes, whereas missense mutations result in mild, late-onset, degenerative phenotypes with limited organ involvement. To test whether this is universally true, in this study we evaluated the genotype–phenotype correlation in 440 individuals from 365 families of an international cohort with NPHP-RC, in whom we detected both recessive mutations in 13 different NPHP genes (NPHP1–NPHP11, AHI1, and CC2D2A). Whereas we had published many of these mutations previously,3,6–11,14–21 in this study, we report nine novel mutations in NPHP2, NPHP3, NPHP4, NPHP5, NPHP6, NPHP8, and AHI1. As for NPHP7 and NPHP9 there were only single families with mutations available, we did not include these genes in our genotype–phenotype study.
RESULTS
Analysis of genotype–phenotype correlation
NPHP1
In 248 individuals from 235 families, the cause of the disease was two recessive NPHP1 mutations, with 93% (219 families) being the homozygous deletion of NPHP1. The patients exhibited a ‘typical’ juvenile NPHP (ESRD ≤4 years) and most of them (180/235 families, 76.5%) had isolated kidney involvement. In 55 families, extrarenal manifestations were noted: specifically, 24 had central nervous system (CNS) defects (oculomotor apraxia-type Cogan, mental retardation, locomotive disorders, CVA, developmental delay, porencephaly, hydrocephalus, and epileptic seizure) and 27 had eye involvement (tapeto-retinal degeneration (TRD), optic nerve atrophy, and pigmented fundus). Two families exhibited liver fibrosis and two had heart anomalies (hypertrophic non-obstructive cardiomyopathy and aortic stenosis).
NPHP2/INVS
In our cohort of 12 families (18 patients) with recessive NPHP2 mutations, 10 carried two nonsense mutations, 1 had two missense mutations, and 1 was compound heterozygous for one nonsense and one missense mutation (Supplementary Table S1 online). The renal phenotype always showed infantile ESRD (at ≥4 years) regardless of the mutation type. In contrast to other forms of NPHP, the patients exhibited enlarged (dysplastic) cystic kidneys. In addition, the phenotypes of the patients with two nonsense mutations were not comparable with each other as they varied from isolated NPHP to JBTS. Extrarenal manifestations were present in 5 of 12 families and included dysplastic phenotypes of the eye (optic nerve atrophy), CNS (hydrocephalus), and the heart (situs inversus, ventricular septal defect, and aortic coarctation). We had one family each with a degenerative phenotype of the eye (TRD) and the heart (hypertrophic cardiomyopathy), and one family with the unusual phenotype of asthma.
NPHP3
Among our cohort of eight families (13 patients) with recessive NPHP3 mutations, four carried two nonsense alleles, two were homozygous for splice site mutations, one was compound heterozygous for one nonsense and one missense allele, and one carried one nonsense and one splice site allele (Supplementary Table S2 online). The renal phenotype always showed ESRD onset between 3 and 13 years regardless of the mutation type, except A1633 (MKS) who passed away at day 3. Extrarenal manifestations were found in four of eight families and included dysplastic phenotypes of the CNS (CVH, molar tooth sign (MTS), and mental retardation) and the heart (atrial septal defect, pulmonary hypoplasia, situs ambiguous, and pulmonary valve stenosis). Two families, with one nonsense mutation and one missense or splice site mutation, had the degenerative phenotype of liver fibrosis.
NPHP4
Among our cohort of 22 families (42 patients) with recessive NPHP4 mutations, 16 carried two nonsense alleles, two had two missense alleles, one was homozygous for a splice site mutation, two were compound heterozygous for one nonsense and one missense mutation, and one was compound heterozygous for one nonsense and one splice site allele (Supplementary Table S3 online). The renal phenotype always showed juvenile ESRD regardless of the mutation type. Interestingly, in two families (one homozygous for a nonsense mutation and the other compound heterozygous for missense mutations) enlarged kidneys were observed. Extrarenal manifestations were present in 10 of 22 families and included dysplastic phenotypes of the eye (coloboma and Leber's congenital amaurosis) and the CNS (mental retardation and developmental delay). There were six families with degenerative phenotypes of the eye (TRD and papillorenal syndrome) and the liver (liver fibrosis), and three with deafness or hearing impairment. There were also the unusual phenotypes of splenomegaly and chronic bronchitis. Interestingly, eight families with two nonsense mutations displayed renal manifestations only.
NPHP5/IQCB1
In our cohort of 25 families (33 patients) with recessive NPHP5 mutations, 23 had two nonsense mutations and two were homozygous for obligatory splice site mutations (Supplementary Table S4 online). Interestingly, despite having two null mutations, the phenotypes of the patient never showed dysplasia in any organ; rather, they always had degenerative phenotypes involving both kidneys and eyes. The renal phenotype showed juvenile ESRD and was always associated with early-onset retinal degeneration (<4 years). One patient had epileptic seizure and one exhibited syndactyly. There was also the unusual phenotype of asthma and one patient had carcinoma in the right kidney and a tumor in the left ovary.
NPHP6/CEP290
Among 19 families (26 patients) with recessive NPHP6 mutations, 14 carried two nonsense alleles, 1 had two missense alleles, 2 were homozygous for obligatory splice site mutations, and 2 were compound heterozygous for one nonsense and one missense or splice site mutation, respectively (Supplementary Table S5 online). All patients developed juvenile ESRD, with two exceptions of infantile ESRD; two patients had enlarged kidneys. All patients with NPHP6 mutations showed extrarenal pheno-types, which included dysplastic phenotypes of the eye (optic nerve atrophy, Leber's congenital amaurosis, and coloboma), the CNS (CVA, CVH, mental retardation, microcephaly, hydrocephalus, and encephalocele occipital), and degenerative phenotypes such as TRD, liver fibrosis, and epileptic seizure.
NPHP8/RPGRIP1L
In eight families (12 patients) with recessive NPHP8 mutations, five had two missense mutations and three were compound heterozygous for one nonsense and one missense mutation (Supplementary Table S6 online). We never found any patient with two nonsense mutations. The renal phenotype always showed juvenile onset of ESRD regardless of the mutation type. Six families had dysplastic phenotypes of the CNS (Dandy–Walker malformation, CVA, developmental delay, encephalocele occipital, MTS, strabismus, and ptosis), four had oculomotor apraxia-type Cogan, and there were two cases with polydactyly, which is a cardinal symptom of patients with Bardet–Biedl syndrome but very rarely occurs in NPHP-RC.
NPHP10/SDCCAG8
In seven families (12 patients) with recessive NPHP10 mutations, five carried two nonsense mutations, one was homozygous for an obligatory splice site mutation, and one was compound heterozygous for a nonsense and a splice site allele (Supplementary Table S7 online). Similar to NPHP5, the phenotypes of patients with NPHP10 mutations were always degenerative involving both kidneys and eyes, but occasionally additional extrarenal manifestations were present. Regardless of the mutation type, all patients in this cohort had juvenile ESRD. Four patients presented with extrarenal manifestations including degenerative phenotype of the CNS (mental retardation, peripheral neuropathy, and cystic brain lesion), generalized seizures, Bardet–Biedl syndrome-like obesity, and hypogenitalism. One patient showed the unusual phenotype of polycystic ovary syndrome.
NPHP11/TMEM67/MKS3
In our 20 families (26 patients) with recessive NPHP11 mutations, 12 had two missense mutations, 6 were compound heterozygous for one nonsense and one missense mutation, and 2 carried one splice site and one missense mutations (Supplementary Table S8 online). In NPHP11, we never detected any patient with two nonsense mutations. Almost all patients exhibited juvenile NPHP, except F278 who exhibited infantile ESRD. In our cohort, all patients exhibited extrarenal manifestations, which mostly included dysplastic phenotypes of the CNS (CVH, CVA, brain atrophy, cerebellar hypoplasia, Dandy–Walker malformation, developmental delay, MTS, oculocerebral disease, mental retardation, strabismus, psychomotor retardation, and ptosis) and the eye (coloboma and optic nerve atrophy), and degenerative liver phenotypes (liver fibrosis, cholangiopathy, and hepatomegaly). Three patients had TRD, one had dysplastic heart phenotype (subvalvular aortic stenosis), and one had dysplastic liver phenotype (cholangiodysplasia). There were also the unusual phenotypes of splenomegaly and Ehlers–Danlos syndrome, and one patient had polydactyly.
AHI1
In our cohort of six families (seven patients) with recessive AHI1 mutations, five had two nonsense mutations and one was homozygous for a splice site mutation (Supplementary Table S9 online). All patients had juvenile NPHP, except F400 who did not have any renal involvement. Extrarenal manifestations were present in all cases and included dysplastic phenotypes of the CNS (MTS and psychomotor developmental delay) and the eye (coloboma). Three patients had TRD, two had tachypnea, and ataxia was reported in one patient.
CC2D2A
In our cohort of three families (three patients) with recessive CC2D2A mutations, all patients were compound heterozygous for one nonsense and one missense mutation (Supplementary Table S10 online). We never detected any patient with two nonsense mutations, although two of three patients had been diagnosed with MKS. All patients showed dysplastic renal phenotypes, one with infantile NPHP. Extrarenal manifestations were present in all patients and included dysplastic phenotypes of the CNS (CVH, mental retardation, and encephalocele occipital), the liver (hepatic developmental defects), and the bone (disarray of bone–cartilage junctions).
Identification of novel mutations in NPHP2, NPHP3, NPHP4, NPHP5, NPHP6, NPHP8, and AHI1
Two patients (families A1423 and A2035) with infantile NPHP (ESRD 12 and 19 months, respectively) were homozygous for a novel nonsense mutation c.2716G>T (p.E906X) in NPHP2.
In two patients, two novel mutations were detected in NPHP3: A1633, who passed away at day 3, had a homozygous frameshift mutation c.3702delA (p.K1234fsX1246), and A1937 (ESRD 8 years 7 months) was homozygous for p.3235C>T (p.Q1079X). Notably, A1633 exhibited an MKS-like phenotype, characterized by CVH, atrial septal defect, double-outlet right ventricle, pulmonary valve stenosis, pulmonary hypoplasia, asplenia, situs ambiguous, and congenital contractures, whereas A1937 showed characteristic features of JBTS including MTS, CVH, and mental retardation.
One novel NPHP4 mutation was detected in A2324 (ESRD 7 years). The patient was homozygous for c.2618_2619insA (p.H873fsX886) and had mental retardation. Interestingly, one sibling with this mutation exhibited TRD and was diagnosed with oculomotor apraxia-type Cogan.
Two novel NPHP5 mutations were detected in two patients with SLSN. F353 was found to be homozygous for c.1363C>T (p.R455X) and A1538 was homozygous for the obligatory splice site mutation c.1130-1G>C. Interestingly, one sibling with the splice site mutation also had syndactyly.
For NPHP6, NPHP8, and AHI1, one novel mutation was identified in each case. A2182 (juvenile ESRD) was homozygous for the obligatory splice site mutation c.2367 + 1G>T in NPHP6 and displayed TRD, mental retardation, and microcephaly. A1685, with two affected siblings (ESRD 15 and 17 years), was homozygous for a missense mutation c.2450A>G (p.Y817C) in NPHP8 and had mental retardation and oculomotor apraxia-type Cogan. A2045 (ESRD 17 years) carried a homozygous deletion c.2172delA (p.I724fsX729) in AHI1.
DISCUSSION
In this study, we have evaluated the genotype–phenotype correlation in a cohort of 440 patients from 365 families with NPHP-RC in whom both mutations were identified. The effect of different NPHP-RC genes on ‘disease phenotype’ is summarized in Table 1, and Figure 1 represents images that are descriptive of the pathological features. As for all patients, mutation screening was not done for all NPHP genes, and thus we cannot conclude the influence of the modifier genes on a phenotype. Mental retardation has been marked as ‘dysplastic’ in case of JBTS patients, who are known to have congenital dysplasia of CNS, and as ‘degenerative’ in patients, who did not have any dysplasia in the brain or any other organ and rather showed degenerative phenotypes only. We observed that a genotype of two null alleles causes phenotypes in the following increasing order of severity: NPHP1, NPHP4, NPHP5, NPHP2, NPHP10, NPHP6 to AHI1. For NPHP8 and NPHP11, no patient was homozygous for two null mutations, most likely because our cohort did not contain a significant number of MKS patients.
Table 1.
Summary of ‘genic’ influences on phenotypes in terms of organ involvement
| Gene (families) | Kidney | Eye | CNS | Liver | Heart | |||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2 null alleles | <2 null alleles | 2 null alleles | <2 null alleles | 2 null alleles | <2 null alleles | 2 null alleles | <2 null alleles | 2 null alleles | <2 null alleles | |
| NPHP1 (226) | 1 (0.4%) + 219 (96.9%) | 6 (2.6%) | 3 (1.3%) + 20 (8.8%) | 1 (0.4%) | 15 (6.6%) + 7 (3%) | 1 (0.4%) | 2 (0.8%) | – | 2 (0.8%) | – |
| NPHP2 (12) | 10 (83.3%) | 2 (16.6%) | 1 (8.3%) + 1 (8.3%) | – | 1 (8.3%) | – | – | – | 2 (16.6%) + 1 (8.3%) | – |
| NPHP3 (5) | 1 (20%) + 3 (60%) | 1 (20%) | – | – | 2 (40%) | – | – | 1 (20%) | 1 (20%) | – |
| NPHP4 (20) | 16 (80%) | 4 (20%) | 3 (15%) + 3 (15%) | 1 (5%) | 2 (10%) | – | 1 (5%) | 1 (5%) | – | – |
| NPHP5 (23) | 23 (100%) | – | 1 (4.3%) + 22 (95.6%) | – | – | – | – | – | – | – |
| NPHP6 (16) | 1 (6.2%) + 13 (81.2%) | 1 (6.2%) + 1 (6.2%) | 8 (50%) + 4 (25%) | 1 (6.2%) + 1 (6.2%) | 11 (68.7%) | 1 (6.2%) | 1 (6.2%) | – | – | – |
| NPHP8 (8) | – | 2 (25%) + 5 (62.5%) | – | – | – | 6 (75%) | – | 1 (12.5%) | – | – |
| NPHP10 (5) | 5 (100%) | – | 4 (80%) | – | 1 (20%) | – | – | – | – | – |
| NPHP11 (18) | – | 1 (5.5%) + 17 (94.4%) | – | 7 (38.5%) + 2 (11.1%) | – | 16 (88%) | – | 1 (5.5%) + 13 (71.5%) | – | 1 (5.5%) |
| AHI1 (5) | 1 (20%) 3 (60%) | – | 1(20%) + 2(40%) | – | 4 (80%) | – | – | – | – | – |
| CC2D2A (3) | – | 3 (100%) | – | – | – | 3 (100%) | – | – | – | 1 (33.3%) |
Abbreviation: CNS, central nervous system.
As splice site mutations can have varying effects, families with splice site mutations were not included in the table. Numbers of patients with dysplastic or degenerative phenotypes are marked with red or green fonts, respectively.
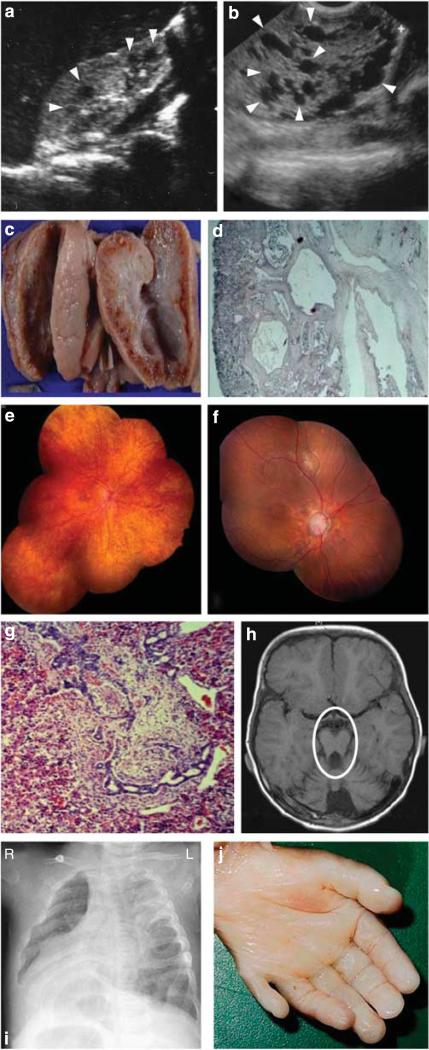
Figure 1. Representative images of degenerative and dysplastic clinical features observed in nephronophthisis (NPHP)-related ciliopathies.
(a) Renal ultrasound from an individual with (degenerative) NPHP. Cysts occur only at the corticomedullary junction and are marked with arrowheads. (b) Renal ultrasound from an individual with (dysplastic) Meckel–Gruber syndrome (MKS, A1633-22). Cysts occur throughout an enlarged kidney and are marked with arrowheads. (c) Autopsy kidney specimen from a fetus with MKS showing bilateral enlarged kidneys interspersed with small, pinhead-sized cysts. (d) Renal histology showing cystic dysplastic kidneys with marked interstitial fibrosis in MKS. (e) Ophthalmoscopy reveals retinitis pigmentosa in the right eye of a patient with (degenerative) Senior–Løken syndrome. (f) Left eye of an NPHP patient with diffuse retinal atrophy with markedly pale discs and enlarged cup. (g) Liver histology showing dysgenesis of the hepatic portal triad with hyperplastic biliary ducts and congenital hepatic fibrosis. (h) Brain magnetic resonance imaging with ‘molar tooth sign’ (marked with a circle) demonstrating vermis hypoplasia, a dysplastic phenotype observed in a JBTS patient. (i) Chest X-ray image of a patient showing situs inversus. (j) Postaxial hexadactyly in an MKS fetus.
Deleterious recessive mutations in NPHP1 caused the mildest disease phenotype compared with the null mutations in other known NPHP-causing genes, resulting mostly in isolated kidney disease. However, about 23.4% (55/235 families) of these patients present with extrarenal organ manifestations, some of which are even dysplastic. We speculate that additional mutations in NPHP-related ‘modifier’ genes might contribute to the development of these extrarenal symptoms as proposed previously.22
In our cohort, all patients with NPHP2 mutations showed ‘infantile ESRD’ (i.e., onset <4 years) with a dysplastic kidney phenotype regardless of the mutation type (i.e., nonsense/missense); however, not all the patients with homozygous null mutations showed extrarenal manifestations, which corroborate the previous observations.10,19,23,24 We conclude that NPHP2 mutations result in ‘infantile NPHP’ with rare extrarenal manifestations (Supplementary Table S1 online). According to Tory et al,24 sometimes different phenotypes were noted for the same mutation, which indicates the possibility of the presence of additional mutations in modifier genes.
No genotype–phenotype correlation was evident for NPHP3 and NPHP4 (Supplementary Tables S2, S3 online), as the phenotypes of the families with two nonsense mutations were not similar. For NPHP3, the phenotypes of A1633 (MKS) and A1937 (JBTS) were more severe compared with F86 and A14 (isolated NPHP). In contrast to some earlier reports,24,25 we did not observe an MKS-like phenotype in all patients with complete loss of NPHP3 function (two null mutations). We conclude that NPHP3 mutations can have diverse effects ranging from infantile ESRD and severe extrarenal manifestations to juvenile ESRD with isolated kidney phenotype. Notably, in our cohort, dysplastic extrarenal manifestations were seen only in patients with two nonsense (null) mutations. For NPHP4, extrarenal manifestations were not always present and we conclude that NPHP4 mutations always result in juvenile ESRD with frequent (7 of 22 families, ~32%) eye involvement.
NPHP5 mutations always resulted in dimorphic disease (SLSN) with juvenile onset NPHP and infantile onset of TRD. Hence, this gene can be stated as a ‘classic’ SLSN-causing gene (Supplementary Table S4 online). Similarly, NPHP10 mutations also resulted in SLSN in most of the patients, often involving extrarenal manifestations in addition (Supplementary Table S7 online).
In our cohort, 17 of 19 families (~90%) with NPHP6 mutations had juvenile NPHP with extrarenal manifestations, and for this gene we found strong allelic influence on the phenotypes (Supplementary Table S5 online). In the families with less than two null alleles, none of the patients showed any dysplastic phenotype and almost never more than two organs were involved. In contrast, in patients with two null mutations always more than two organs were involved, and the patients showed at least one dysplastic phenotype. Notably, in the previous reports, the genotype–phenotype correlation was not clear for NPHP6, as phenotypes of the patients with two null mutations varied from MKS to JBTS to Leber's congenital amaurosis.14,26,27 We speculate that the reason for this observation might be the fact that the domain of the protein being mutated is crucial.
According to the literature, complete loss of function of NPHP8 leads to MKS, and two missense mutations lead to a degenerative phenotype of kidneys and eyes.3 We have one MKS patient in our cohort, but the patient is compound heterozygous for a nonsense and a missense mutation. Hence, we conclude that NPHP8 mutations cause severe phenotypes irrespective of the mutation type, because in our cohort at least six of eight families have dysplastic phenotypes of the CNS (Supplementary Table S6). Similarly, any NPHP11 mutation leads to severe phenotypes (Supplementary Table S8). In our cohort, patients with two missense mutations in NPHP11 almost always developed liver fibrosis (16 of 20 families), CVA/CVH (10 of 20 families), and mental retardation (14 in 20 families). In addition, retinal coloboma was present in 8 families.
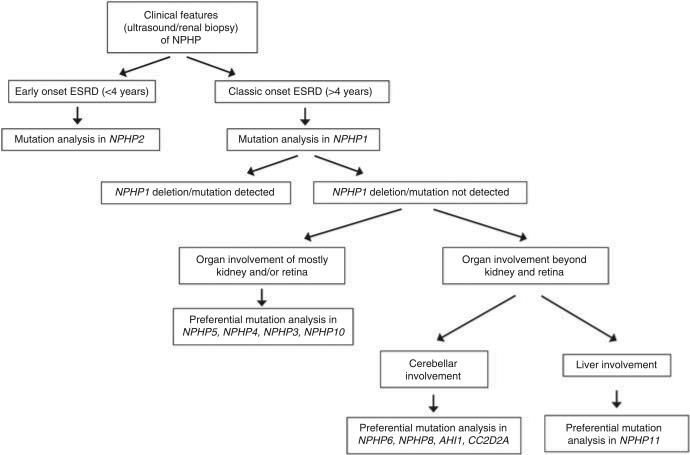
For AHI1, we observed that all our patients with two nonsense mutations have JBTS (Supplementary Table S9 online), which is concordant with other reports.28–30 Therefore, AHI1 can be classified as a ‘JBTS gene’, as all the patients with mutations in this gene exhibit JBTS with the pathognomonic feature of the ‘molar tooth sign’ seen by magnetic resonance imaging. However, for CC2D2A, the number of patients is too small to draw any conclusion on the genotype–phenotype correlation. In our cohort of three patients, CC2D2A mutations always lead to severe pheno-types; therefore, it is most likely that CC2D2A is an MKS or JBTS gene rather than an isolated NPHP gene. Thus, according to the data obtained from our cohort, we suggest a genetic analysis pathway based on the presence of frequent and less frequent phenotypes (Figure 2). We conclude that the ‘gene locus heterogeneity’ is the prime factor in determining the disease phenotype, whereas ‘allelism’ might be somewhat important only for certain genes such as NPHP6, NPHP8, and NPHP11.
Figure 2.
Flowchart for a suggested genotyping strategy in nephronophthisis (NPHP)-related ciliopathies (Meckel–Gruber syndrome patients are excluded). ESRD, end-stage renal disease.
MATERIALS AND METHODS
Patients
We collected blood samples, pedigrees, and clinical information with informed consent (http://www.renalgenes.org) from a worldwide cohort of 365 families with MKS, JBTS, SLSN, or isolated NPHP. Approval for studies on human subjects was obtained from the University of Michigan Institutional Review Board. In all patients, the diagnosis of NPHP was based on one or more of the following criteria: (i) clinical course with characteristic clinical signs of polyuria, polydipsia, anemia, and growth retardation; (ii) presence of chronic renal failure; (iii) renal ultrasound or renal biopsy compatible with the diagnosis of NPHP as judged by a pediatric nephrologist; (iv) pedigree compatible with autosomal recessive inheritance. Neurological criteria for JBTS were based on the following clinical hallmarks of this cerebello-oculo-renal syndrome: MTS or diagnosis of JBTS by a neurologist (pediatric) or geneticist. In addition, associated JBTS symptoms were recorded: nervus opticus or retinal coloboma, TRD, CVA/CVH, ataxia, and periodic apnea/tachypnea. The diagnosis of SLSN was based on the presence of NPHP in association with TRD.
Mutation analysis of NPHP1–NPHP11, AHI1, and CC2D2A
Genomic DNA from peripheral blood samples was extracted by standard methods. Mutations were detected by homozygosity mapping31 followed by PCR amplification of respective exons with flanking primers and direct sequencing of each amplicon using the dideoxy chain termination method on an Applied Biosystems (ABI) capillary sequencer (Applied Biosystems). Resulting sequences were aligned and evaluated by Sequencher (version 3.8) software (Gene Codes Corporation, Ann Arbor, MI). Novel mutations were found to be absent from at least 90 ethnically matched healthy controls.
Supplementary Material
Supplementary Table S1: Genotype-phenotype correlation in 12 families with two recessive NPHP2/INVS mutations1
Supplementary Table S2: Genotype-phenotype correlation in 8 families with two recessive NPHP3 mutations1
Supplementary Table S3: Genotype-phenotype correlation in 22 families with two recessive NPHP4 mutations1
Supplementary Table S4: Genotype-phenotype correlation in 25 families with two recessive NPHP5/IQCB1 mutations1
Supplementary Table S5: Genotype-phenotype correlation in 19 families with two recessive NPHP6/CEP290 mutations1
Supplementary Table S6: Genotype-phenotype correlation in 8 families with two recessive NPHP8/RPGRIP1L mutations1
Supplementary Table S7: Genotype-phenotype correlation in 7 families with two recessive NPHP10/SDCCAG8 mutations1
Supplementary Table S8: Genotype-phenotype correlation in 20 families with two recessive NPHP11/TMEM67/MKS3 mutations1
Supplementary Table S9: Genotype-phenotype correlation in 6 families with two recessive AHI1 mutations1
Supplementary Table S10: Genotype-phenotype correlation in 3 families with two recessive CC2D2A mutations1
ACKNOWLEDGMENTS
We thank Kari Branham for providing patient clinical data. We sincerely thank the affected individuals and their families for participation. We also thank the physicians who contributed to this study. We acknowledge RH Lyons for excellent next-generation sequencing. FH is an Investigator of the Howard Hughes Medical Institute, a Doris Duke Distinguished Clinical Scientist, and a Frederick G L Huetwell Professor. This research was supported by grants from the National Institutes of Health to FH (DK068306 and DK090947).
Footnotes
DISCLOSURE
All the authors declared no competing interests.
REFERENCES
- 1.Hildebrandt F, Sayer JA, Jungers P, et al. Nephronophthisis-medullary cystic and medullary sponge kidney disease. In: Schrier RW, editor. Diseases of the Kidney and Urinary Tract. 8th edn Vol. 1. Wolters kluwer health/Lippincott Williams & Wilkins; Philadelphia: 2007. pp. 478–501. [Google Scholar]
- 2.Attanasio M, Uhlenhaut NH, Sousa VH, et al. Loss of GLIS2 causes nephronophthisis in humans and mice by increased apoptosis and fibrosis. Nat Genet. 2007;39:1018–1024. doi: 10.1038/ng2072. [DOI] [PubMed] [Google Scholar]
- 3.Delous M, Baala L, Salomon R, et al. The ciliary gene RPGRIP1L is mutated in cerebello-oculo-renal syndrome (Joubert syndrome type B) and Meckel syndrome. Nat Genet. 2007;39:875–881. doi: 10.1038/ng2039. [DOI] [PubMed] [Google Scholar]
- 4.Gorden NT, Arts HH, Parisi MA, et al. CC2D2A is mutated in Joubert syndrome and interacts with the ciliopathy-associated basal body protein CEP290. Am J Hum Genet. 2008;83:559–571. doi: 10.1016/j.ajhg.2008.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hildebrandt F, Otto E, Rensing C, et al. A novel gene encoding an SH3 domain protein is mutated in nephronophthisis type 1. Nat Genet. 1997;17:149–153. doi: 10.1038/ng1097-149. [DOI] [PubMed] [Google Scholar]
- 6.Olbrich H, Fliegauf M, Hoefele J, et al. Mutations in a novel gene, NPHP3, cause adolescent nephronophthisis, tapeto-retinal degeneration and hepatic fibrosis. Nat Genet. 2003;34:455–459. doi: 10.1038/ng1216. [DOI] [PubMed] [Google Scholar]
- 7.Otto E, Loeys B, Khanna H, et al. A novel ciliary IQ domain protein, NPHP5, is mutated in Senior-Loken syndrome (nephronophthisis with retinitis pigmentosa), and interacts with RPGR and calmodulin. Nat Genet. 2005;37:282–288. doi: 10.1038/ng1520. [DOI] [PubMed] [Google Scholar]
- 8.Otto E, Hoefele J, Ruf R, et al. A gene mutated in nephronophthisis and retinitis pigmentosa encodes a novel protein, nephroretinin, conserved in evolution. Am J Hum Genet. 2002;71:1167–1171. doi: 10.1086/344395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otto EA, Hurd TW, Airik R, et al. Candidate exome capture identifies mutation of SDCCAG8 as the cause of a retinal-renal ciliopathy. Nat Genet. 2010;42:840–850. doi: 10.1038/ng.662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Otto EA, Schermer B, Obara T, et al. Mutations in INVS encoding inversin cause nephronophthisis type 2, linking renal cystic disease to the function of primary cilia and left-right axis determination. Nat Genet. 2003;34:413–420. doi: 10.1038/ng1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Otto EA, Tory K, Attanasio M, et al. Hypomorphic mutations in meckelin (MKS3/TMEM67) cause nephronophthisis with liver fibrosis (NPHP11). J Med Genet. 2009;46:663–670. doi: 10.1136/jmg.2009.066613. [DOI] [PubMed] [Google Scholar]
- 12.Otto EA, Trapp ML, Schultheiss UT, et al. NEK8 mutations affect ciliary and centrosomal localization and may cause nephronophthisis. J Am Soc Nephrol. 2008;19:587–592. doi: 10.1681/ASN.2007040490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saunier S, Calado J, Benessy F, et al. Characterization of the NPHP1 locus: mutational mechanism involved in deletions in familial juvenile nephronophthisis. Am J Hum Genet. 2000;66:778–789. doi: 10.1086/302819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Sayer JA, Otto EA, O'Toole JF, et al. The centrosomal protein nephrocystin-6 is mutated in Joubert syndrome and activates transcription factor ATF4. Nat Genet. 2006;38:674–681. doi: 10.1038/ng1786. [DOI] [PubMed] [Google Scholar]
- 15.Utsch B, Sayer JA, Attanasio M, et al. Identification of the first AHI1 gene mutations in nephronophthisis-associated Joubert syndrome. Pediatr Nephrol. 2006;21:32–35. doi: 10.1007/s00467-005-2054-y. [DOI] [PubMed] [Google Scholar]
- 16.Helou J, Otto EA, Attanasio M, et al. Mutation analysis of NPHP6/CEP290 in patients with Joubert syndrome and Senior-Loken syndrome. J Med Genet. 2007;44:657–663. doi: 10.1136/jmg.2007.052027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hoefele J, Sudbrak R, Reinhardt R, et al. Mutational analysis of the NPHP4 gene in 250 patients with nephronophthisis. Hum Mutat. 2005;25:411. doi: 10.1002/humu.9326. [DOI] [PubMed] [Google Scholar]
- 18.Hoefele J, Wolf MT, O'Toole JF, et al. Evidence of oligogenic inheritance in nephronophthisis. J Am Soc Nephrol. 2007;18:2789–2795. doi: 10.1681/ASN.2007020243. [DOI] [PubMed] [Google Scholar]
- 19.Otto EA, Helou J, Allen SJ, et al. Mutation analysis in nephronophthisis using a combined approach of homozygosity mapping, CEL I endonuclease cleavage, and direct sequencing. Hum Mutat. 2008;29:418–426. doi: 10.1002/humu.20669. [DOI] [PubMed] [Google Scholar]
- 20.Otto EA, Ramaswami G, Janssen S, et al. Mutation analysis of 18 nephronophthisis associated ciliopathy disease genes using a DNA pooling and next generation sequencing strategy. J Med Genet. 2011;48:105–116. doi: 10.1136/jmg.2010.082552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wolf MT, Saunier S, O'Toole JF, et al. Mutational analysis of the RPGRIP1 L gene in patients with Joubert syndrome and nephronophthisis. Kidney Int. 2007;72:1520–1526. doi: 10.1038/sj.ki.5002630. [DOI] [PubMed] [Google Scholar]
- 22.Tory K, Lacoste T, Burglen L, et al. High NPHP1 and NPHP6 mutation rate in patients with Joubert Syndrome and nephronophthisis: potential epistatic effect of NPHP6 and AHI1 mutations in patients with NPHP1 mutations. J Am Soc Nephrol. 2007;18:1566–1575. doi: 10.1681/ASN.2006101164. [DOI] [PubMed] [Google Scholar]
- 23.O'Toole JF, Otto EA, Frishberg Y, et al. Retinitis pigmentosa and renal failure in a patient with mutations in INVS. Nephrol Dial Transplant. 2006;21:1989–1991. doi: 10.1093/ndt/gfl088. [DOI] [PubMed] [Google Scholar]
- 24.Tory K, Rousset-Rouviere C, Gubler MC, et al. Mutations of NPHP2 and NPHP3 in infantile nephronophthisis. Kidney Int. 2009;75:839–847. doi: 10.1038/ki.2008.662. [DOI] [PubMed] [Google Scholar]
- 25.Bergmann C, Fliegauf M, Bruchle NO, et al. Loss of nephrocystin-3 function can cause embryonic lethality, Meckel-Gruber-like syndrome, situs inversus, and renal-hepatic-pancreatic dysplasia. Am J Hum Genet. 2008;82:959–970. doi: 10.1016/j.ajhg.2008.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Baala L, Audollent S, Martinovic J, et al. Pleiotropic effects of CEP290 (NPHP6) mutations extend to Meckel syndrome. Am J Hum Genet. 2007;81:170–179. doi: 10.1086/519494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.den Hollander AI, Koenekoop RK, Yzer S, et al. Mutations in the CEP290 (NPHP6) gene are a frequent cause of Leber congenital amaurosis. Am J Hum Genet. 2006;79:556–561. doi: 10.1086/507318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dixon-Salazar T, Silhavy JL, Marsh SE, et al. Mutations in the AHI1 gene, encoding jouberin, cause Joubert syndrome with cortical polymicrogyria. Am J Hum Genet. 2004;75:979–987. doi: 10.1086/425985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ferland RJ, Eyaid W, Collura RV, et al. Abnormal cerebellar development and axonal decussation due to mutations in AHI1 in Joubert syndrome. Nat Genet. 2004;36:1008–1013. doi: 10.1038/ng1419. [DOI] [PubMed] [Google Scholar]
- 30.Kroes HY, van Zon PH, Fransen van de Putte D, et al. DNA analysis of AHI1, NPHP1 and CYCLIN D1 in Joubert syndrome patients from the Netherlands. Eur J Med Genet. 2008;51:24–34. doi: 10.1016/j.ejmg.2007.10.001. [DOI] [PubMed] [Google Scholar]
- 31.Hildebrandt F, Heeringa SF, Ruschendorf F, et al. A systematic approach to mapping recessive disease genes in individuals from outbred populations. PLoS Genet. 2009;5:e1000353. doi: 10.1371/journal.pgen.1000353. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary Table S1: Genotype-phenotype correlation in 12 families with two recessive NPHP2/INVS mutations1
Supplementary Table S2: Genotype-phenotype correlation in 8 families with two recessive NPHP3 mutations1
Supplementary Table S3: Genotype-phenotype correlation in 22 families with two recessive NPHP4 mutations1
Supplementary Table S4: Genotype-phenotype correlation in 25 families with two recessive NPHP5/IQCB1 mutations1
Supplementary Table S5: Genotype-phenotype correlation in 19 families with two recessive NPHP6/CEP290 mutations1
Supplementary Table S6: Genotype-phenotype correlation in 8 families with two recessive NPHP8/RPGRIP1L mutations1
Supplementary Table S7: Genotype-phenotype correlation in 7 families with two recessive NPHP10/SDCCAG8 mutations1
Supplementary Table S8: Genotype-phenotype correlation in 20 families with two recessive NPHP11/TMEM67/MKS3 mutations1
Supplementary Table S9: Genotype-phenotype correlation in 6 families with two recessive AHI1 mutations1
Supplementary Table S10: Genotype-phenotype correlation in 3 families with two recessive CC2D2A mutations1