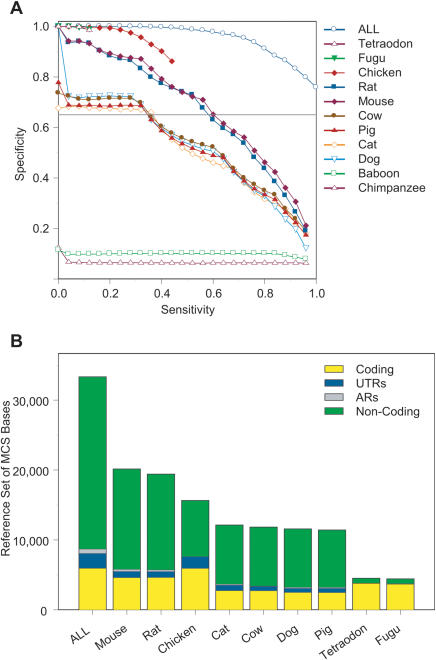
Figure 7.
Ability of individual species' sequences to detect MCSs. (A) Using the indicated species' sequences, MCSs were identified by the binomial-based method over a range of conservation score thresholds. Shown is the resulting relationship between sensitivity (fraction of reference MCS bases detected; see Methods) and specificity (fraction of detected MCS bases that corresponds to reference MCS bases). Also indicated are the results using the sequences from all 11 non-human species (ALL). Note that the limited amount of alignable sequence from chicken and fish impedes the ability to obtain the full range of sensitivity/specificity values. (B) Detection of reference MCS bases, indicated for each type of sequence (coding, UTRs, ARs, and non-coding). This is shown for each species' sequence using the data obtained with a specificity of 65% (horizontal grey line in A), except for chicken and fish. For the latter species, data obtained with specificities of 81% and 99%, respectively, were used (since lower specificities cannot be achieved with these sequences; see A). ALL represents the entire set of reference MCS bases (which is detected by the binomial-based method with a 75% specificity when the sequences from all 11 non-human species are used). Data with non-human primate sequences were not included in B because of their inability to achieve a specificity of 65% (see A). Note that a specificity of 65% was chosen since it allowed the inclusion of most species' sequences. The underlying data associated with these analyses are available at http://www.nisc.nih.gov/data.