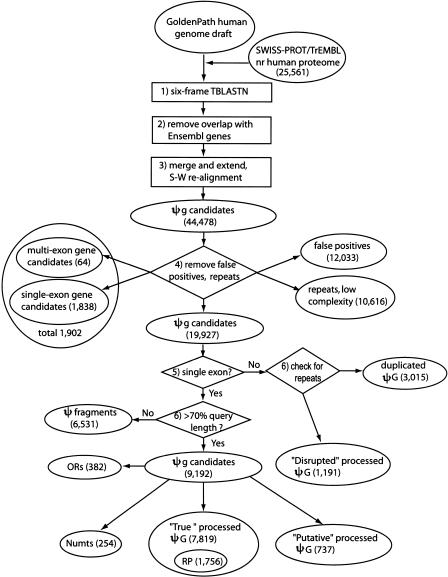
Figure 9.
A flow chart showing procedures in searching for processed pseudogenes in the human genome. (RP) ribosomal proteins; (ΨG) pseudogene; (OR) olfactory receptor; (Numts) nuclear mitochondrial pseudogenes; (S-W) Smith-Waterman. The steps are as follows: (1) Six-frame TBLASTN run searching for SWISS-PROT/TrEMBL protein similarities in the human genome. (2) Remove overlaps with Ensembl functional gene annotations. (3) Merging, extension, and realignment. BLAST hits were merged and extended on both sides to match the length of query protein sequence and then realigned with the protein sequence. After this step, 44,478 pseudogene candidates were obtained. (4) Remove false positives, repeats, low complexity sequences, and potential functional gene candidates. A total of 19,927 pseudogene candidates were obtained at this step. In steps 5 and 6, processed pseudogenes, duplicated pseudogenes, and pseudogene fragments were separated according to sequence continuity and completeness. Two special types of pseudogene, ORs and Numts, were further removed from the pool, and processed pseudogenes were grouped into three classes, “True,” “Putative,” and “Disrupted.” See text for the definition of these three classes.