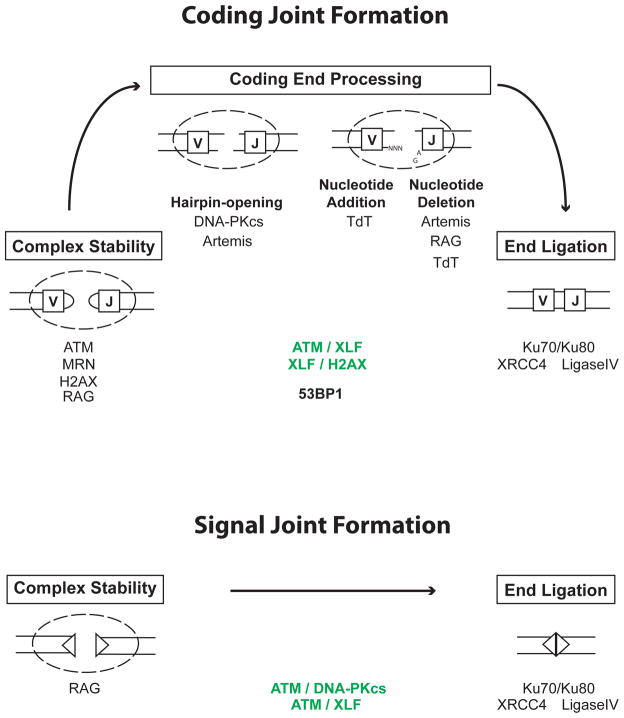
Figure 2. Factors involved in signal and coding joint formation.
RAG cleavage generates a pair of hairpin sealed coding ends (rectangles) and a pair of blunt phosphorylated signal ends (triangles). After RAG cleavage, coding ends and signal ends reside in complexes until they can be processed and joined. Coding end complexes are stabilized by ATM and MRN and may also be stabilized by H2AX and possibly RAG. ATM and MRN do not appear to be critical for signal end complex stability. However, ATM and DNA-PKcs have overlapping activities (shown in green) that are required for signal joint formation. Although the basis for this requirement is unknown, it could reflect the overlapping activity of ATM and DNA-PKcs in promoting signal end complex stability. RAG stays bound to signal ends in post-cleavage complexes generated in vitro and it has been suggested that RAG may stabilize signal ends in vivo, but a direct demonstration of this RAG function is lacking. Ku70/80 and DNA-PKcs, which are recruited early to DSBs, have been implicated in the stabilization of DNA ends prior to joining but their potential function in stabilizing RAG DSBs has not been examined.
Hairpin sealed coding ends are opened through the endonuclease activity of Artemis. Its endonuclease activity requires association with and possibly phosphorylation by activated DNA-PKcs. Nucleotides can then be added to coding ends through the activity of TdT, and nucleotides can be lost through the nuclease activity of Artemis and possibly RAG and TdT. The joining of signal and coding ends is carried out by DNA Ligase IV and XRCC4 with Ku70/Ku80 potentially being important to align DNA ends prior to joining. Although Ku function is essential for both signal and coding joint formation, its specific activities at RAG DSBs have not been directly examined. Notably, ATM and XLF have overlapping activities (shown in green) that are also important for signal and coding joint formation. In addition, XLF has overlapping activities with H2AX that are required for coding joint formation. Whether these overlapping activities are also required for signal joint formation has not been examined. The nature of these overlapping activities of XLF and ATM or H2AX remains to be determined. 53BP1 is required for optimal coding joint formation for reasons that are not clear but may involve a function in long range joining and/or preventing DNA end resection.