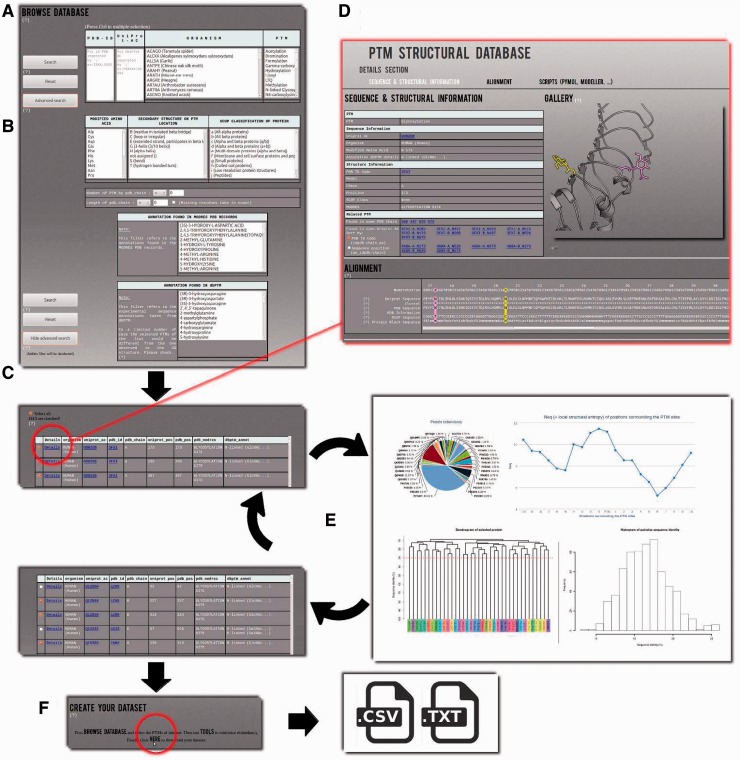
Figure 2.
Example of PTM-SD usage. (A) A simple mode is available to use PTM-SD. It is possible to look for a list of PDB and/or UniProt ids, combined with a specific organism and (a) particular PTM(s). (B) An advanced mode allows more complex requests, as specific amino acid(s), secondary structure(s), SCOP fold(s), number of PTM by PDB chain, length of PDB chain and detailed annotation(s) found in PDB records and dbPTM. By clicking on the search button, (C) a results table appears. It gives for each entry the information on organism, cross-linking with the PDB and UniProt id, the precise position of the PTM in sequence and structural data, PTM annotations and its structural environment in terms of PBs and secondary structures. (D) By clicking on the ‘details’ link, the visitor is redirected to a new page containing extra information on the selected PTM site. Related PTM-SD entries found in same PDB chain and same UniProt AC are accessible through cross-link. Image gallery was done, thanks to PyMOL software, and below alignment section, done thanks to Clustal W, allows direct observation of the sequence/structure relationship surrounding PTM sites. On this page, scripts for PyMOL software and template sequence for Modeller comparative software are provided. (E) From the complete set of selected entries, it is possible to look at the distribution of organisms, proteins, PDB ids/chains and PTM types. Neq entropy index quantifies the local structural divergences between the PTMs. From this step, the visitor can reduce the protein redundancy into his selected entries. (F) At last he can download the list of PDB chain and the PTM-SD data related to his selection.