Figure 4. Hotair regulates silent chromatin state genome-wide.
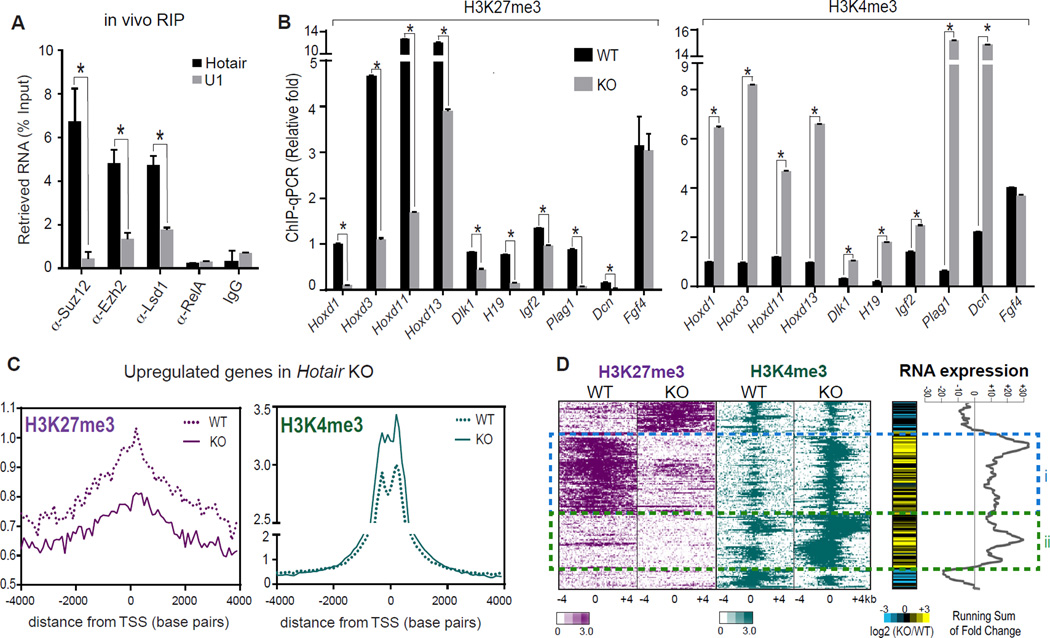
(A) Hotair binds PRC2 and LSD1 complexes in vivo. RIP of E11.5 embryos with the indicated antibodies was followed by qRT-PCR of Hotair and control RNAs (U1, Malat1) and normalized with 1% input performed in parallel (n=3). Mean±s.d. is shown for all panels; *, p<0.05 student's t-test.
(B) ChIP-qPCR in Hotair KO vs. WT cells of H3K27me3 (left panel) and H3K4me3 (right panel) of the indicated genes. Fgf4 was not changed in KO and served as a negative control. (n=3).
(C) Average H3K27me3 (left panel) and H3K4me3 (right panel) ChIP-seq signal across the transcription start site (TSS) of upregulated genes in Hotiar KO cells is shown.
(D) Relationship of histone modifications to gene de-repression in Hotair WT and KO cells. Heat map zoom-in of ChIP-seq signal in 8-kb bins centered on peak summits after unsupervised hierarchical clustering (left). RNA expression changes (KO/WT) and running sums across clusters are shown (right). Gene activation is seen in cluster i (75 loci) with both H3K27me3 loss and H3K4me3 gain; cluster ii shows 70 loci with only H3K4me3 gain in Hotair KO.
See also Figure S4.