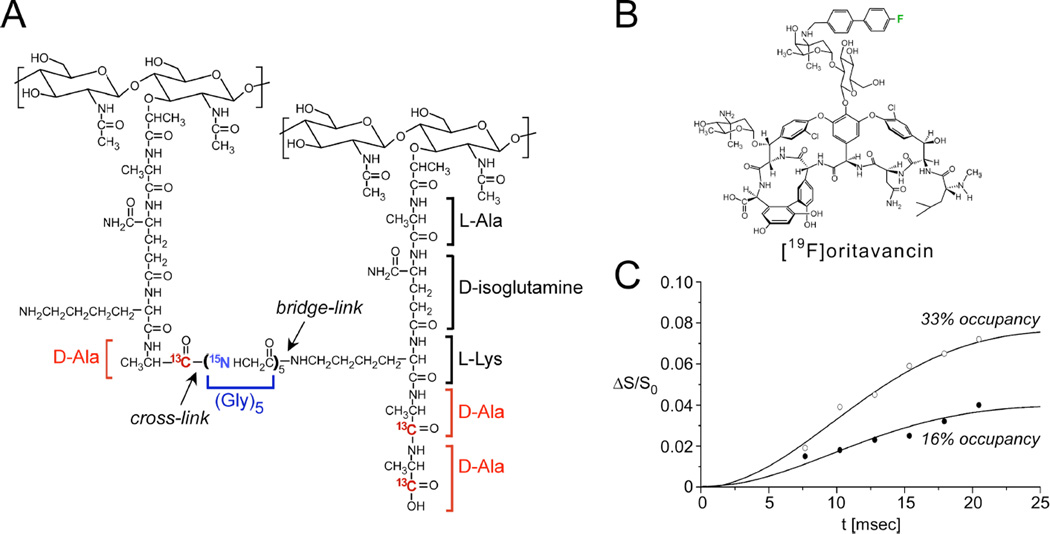
Figure 4. Distances between an antibiotic and the bacterial cell wall.
(A) Chemical schematic of the S. aureus peptidoglycan, highlighting the unique bridge-link and crosslink sites that are often targeted in REDOR experiments through the use of selective D-[1-13C]Ala, [15N]Gly, and L-[ε-15N]Lys labeling. (B) Chemical structure of [19F]oritavancin. (C) 13C{19F} REDOR dephasing (ΔS/S0) as a function of the dipolar evolution time, t, for complexes of [19F]oritavancin with peptidoglycan isolated from S. aureus whole cells grown on media containing D-[1-13C]alanine. The binding site occupancy of [19F]oritavancin complexed to cell walls was either 33% (open circles) or 16% (closed circles), wherein 2 µmol antibiotic was bound to 20 or 40 mg, respectively, of isolated peptidoglycan (dry mass). The calculated cell wall dephasing (solid lines) assumed a Gaussian distribution of distances for isolated 13C-19F pairs centered at 7.6 Å with a width of 1.5 Å. The dashed line also provides a reasonable fit to the data. Spectra were obtained on a 500 MHz spectrometer. Figure adapted from Kim et al.35