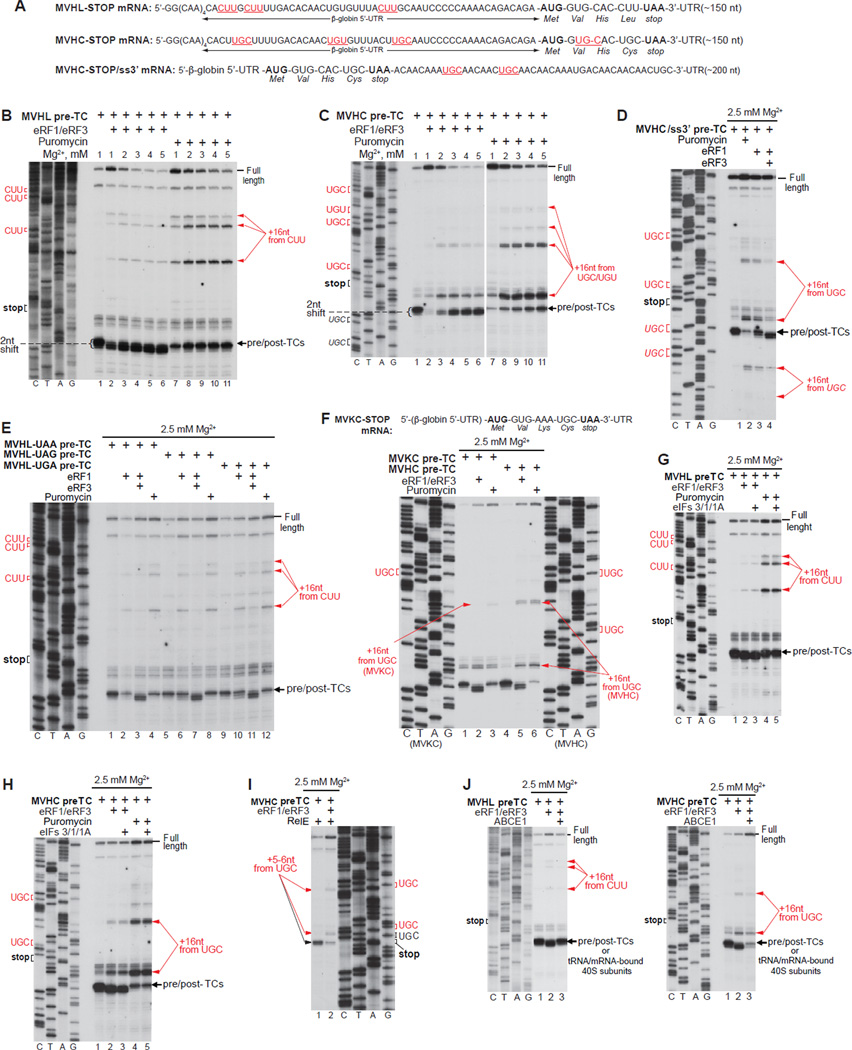
Figure 1. Mammalian post-termination ribosomes can migrate to nearby codons that are cognate to the P-site deacylated tRNA.
(A; F, upper panel) Structure of (A) MVHL-STOP, MVHC-STOP, MVHC-STOP/ss3’ and (F) MVKC-STOP mRNAs. Upstream and downstream codons that are identical to the last codon of the encoded tetrapeptide are in red and underlined (A). (B-H, J) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on MVHL-STOP, MVHC-STOP, MVHC-STOP/ss3’ or MVKC-STOP mRNAs with combinations of eRF1, eRF3, puromycin, eIFs 3/1/1A and ABCE1 at the indicated free [Mg2+]. Black arrows show the positions of pre-TCs, post-TCs and recycled 40S subunits. Red arrows indicate the positions of migrated post-termination ribosomes. Upstream and downstream codons that are identical to the last codon of the encoded tetrapeptide are in red. (I) Primer-extension analysis of A-site mRNA cleavage induced by RelE in pre- and post-TCs assembled on MVHC-STOP mRNA. The positions of cleavages and of corresponding P-site codons are indicated.