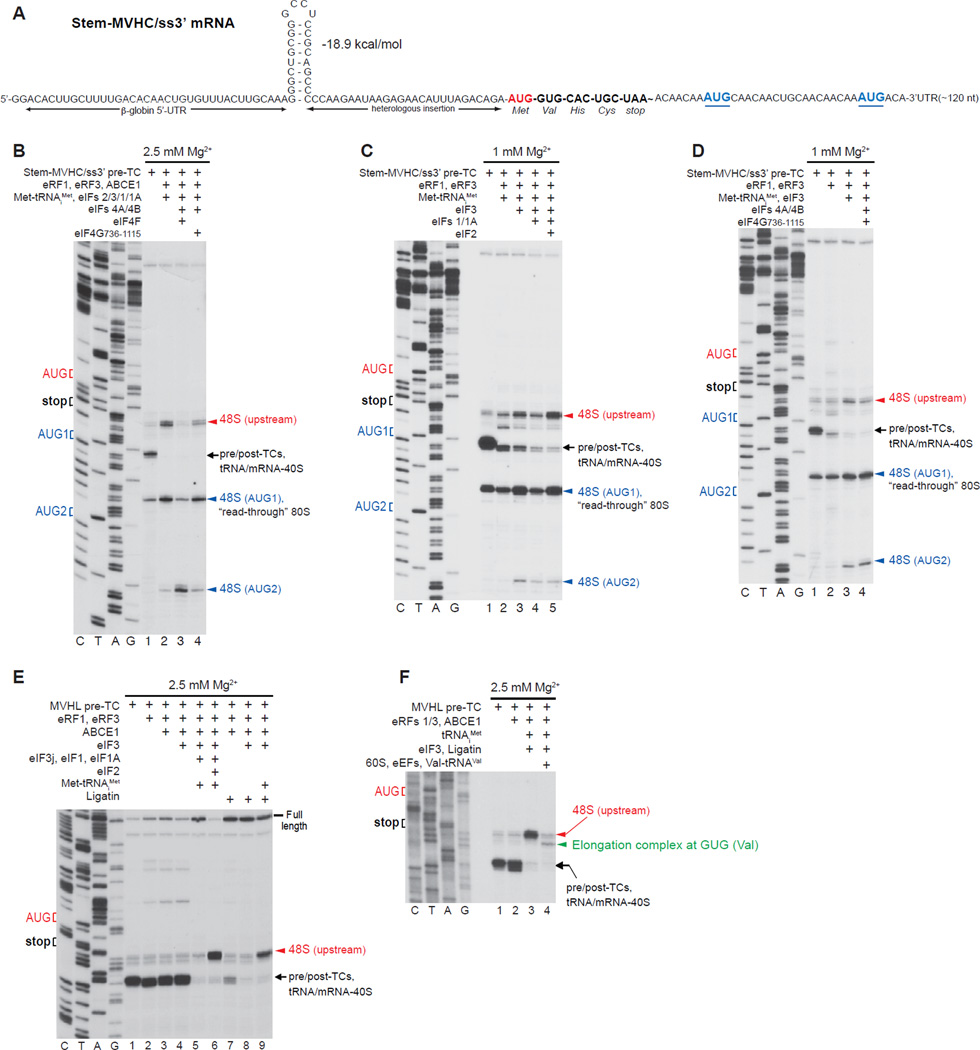
Figure 7. Reinitiation on mRNA with an unstructured region downstream of the stop codon.
(A) Structure of Stem-MVHC/ss3’ mRNA. (B-D) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on Stem-MVHC/ss3’ mRNA with combinations of eRF1, eRF3, ABCE1, Met-tRNAiMet and eIFs 2/3/1/1A/4A/4B/4F/4G at the indicated free [Mg2+]. The positions of pre-TCs, post-TCs and tRNA/mRNA-associated recycled 40S subunits are indicated by black arrows; positions of 48S complexes formed at the AUG triplets upstream and downstream of the stop codon are shown in red and blue, respectively. (E, F) Toe-printing analysis of ribosomal complexes obtained by treating pre-TCs formed on MVHL-STOP mRNA with different combinations of eRF1, eRF3, ABCE1, Met-tRNAiMet, tRNAiMet, eIFs 2/3/1/1A, Ligatin, eEFs, 60S subunits and Val-tRNAVal at the indicated free [Mg2+]. The positions of pre-TCs, post-TCs and tRNA/mRNA-associated recycled 40S subunits are indicated by black arrows; positions of 48S complexes formed at the upstream AUG triplet and to elongation complexes formed at the GUG codon are shown in red and green, respectively.