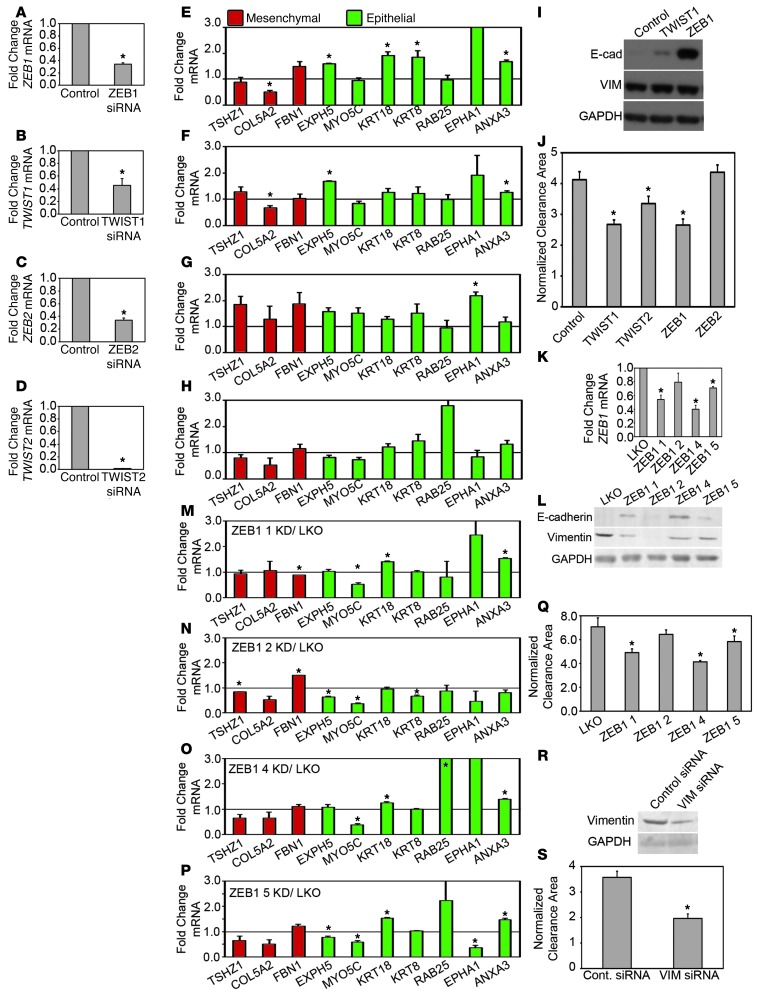
Figure 3. Knockdown of EMT transcription factors or vimentin inhibits mesothelial clearance.
(A–D) qRT-PCR measurements of mRNA levels of (A) ZEB1, (B) TWIST1, (C) ZEB2, and (D) TWIST2 in OVCA433 cells transfected with siRNA SMARTpools targeting luciferase (control), ZEB1, TWIST1, ZEB2, or TWIST2. (E–H) qRT-PCR measurements of mRNA levels of EMT markers in (E) ZEB1, (F) TWIST1, (G) ZEB2, or (H) TWIST2 siRNA–treated OVCA433 cells. Measurements were normalized to RPLPO mRNA levels and expressed as fold changes compared to controls. Data are shown as the mean of 3 biological replicates for each condition. Each biological replicate was derived from an average of 3 technical replicates. (I, L, and R) Western blot analysis of E-cadherin and vimentin in OVCA433 cells transfected with (I) siRNA SMARTpools targeting luciferase, TWIST1, or ZEB1; (L) with empty vector control or shRNAs targeting ZEB1; and (R) with siRNA SMARTpools targeting luciferase or vimentin. (J, Q, and S) Normalized average clearance area of ZT mesothelial monolayers at 8 hours after coincubation with OVCA433 spheroids transfected with (J) luciferase, TWIST1, TWIST2, ZEB1, or ZEB2 siRNA SMARTpools; (Q) empty vector control or shRNAs targeting ZEB1; and (S) siRNA SMARTpools targeting luciferase or vimentin. >60 positions scored per condition in 3 independent experiments. (K) qRT-PCR measurements of mRNA levels of ZEB1 in OVCA433 cells transfected with empty vector control (LKO) or shRNAs targeting ZEB1. (M–P) qRT-PCR measurements of mRNA levels of EMT markers in OVCA433 cells transfected with ZEB1 (M) shRNA 1, (N) shRNA 2, (O) shRNA 4, or (P) shRNA 5 normalized to control marker expression. Error bars denote SEM. *P < 0.05, Student’s t test.