Figure 4.
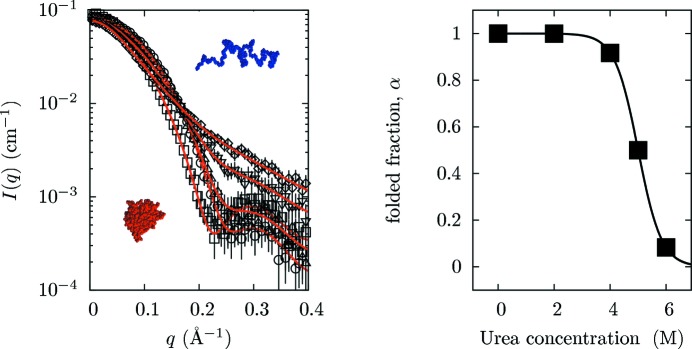
(Left) Simulated SANS curves obtained for BLG in D2O at 5 g l−1 with increasing urea concentration in solution (from bottom to top, open squares, circles, up-triangles, down-triangles and diamonds correspond to 0, 2, 4, 5 and 6 M urea, respectively) and their best fits obtained with GENFIT. All data were simulated at ambient pressure and temperature, at pD = 2.3, and at 20 mM ionic strength. The native BLG monomer and the unfolded chain are reported. The best fit parameters of the worm-like monomer were Kuhn length b = 4.6 ± 0.4 Å, inner cross section R = 4.1 ± 0.2 Å, number of statistical segments N b = 90 ± 20 and relative mass density of the hydration shell d w = 0.951 ± 0.001. The best fit parameters of the unfolding free energy are ΔG unf,0 = 12 ± 1 k B T, ΔG unf,1 = −2.4 ± 0.2 k B T M −1 and ΔG unf,2 = 0.00 ± 0.03 k B T M −2. (Right) BLG folded monomer fraction in solution versus urea molar content as obtained from the calculated unfolding free energy.
