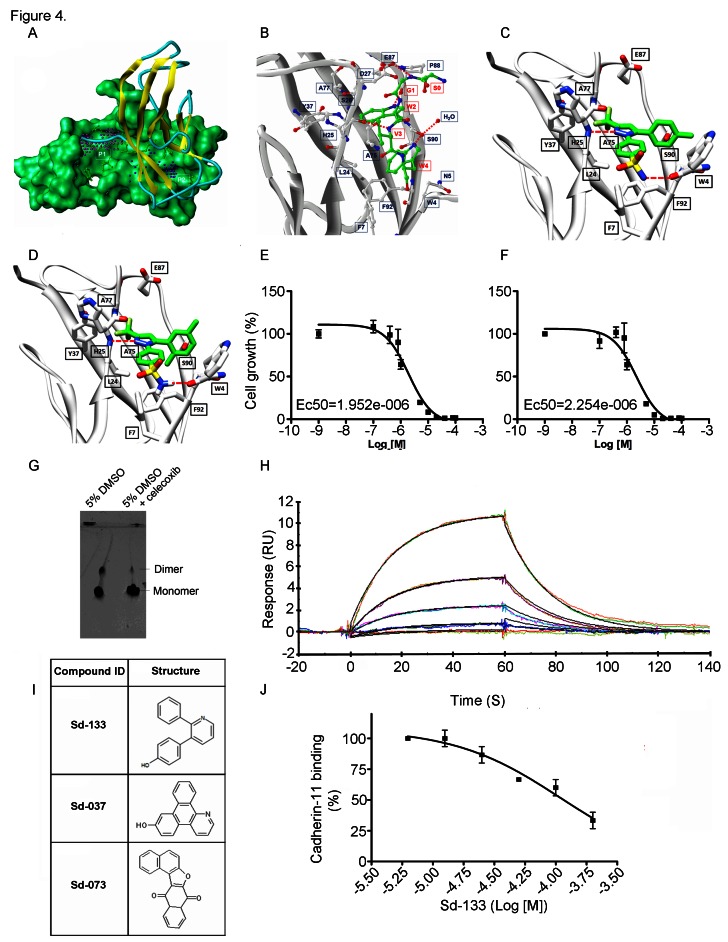
Figure 4. Structural modeling of celecoxib, DMC and other small molecule inhibitors binding to CDH11 and inhibiting the growth of MDA-MB-231 cells. Sd-133 binding capability to CDH11 was validated by SPR.
(A) EC1 homodimer interface of CDH11 (PDB: 2A4C); one monomer is represented by the Van der Waals molecular surface (green) and the other by a ribbon. P1 is a hydrophobic, concave surface binding to two W residues from the partner EC1 monomer. P2 is a small pocket defined by the EC1 domain itself. Virtual screening was carried out with the residues lining P1 and P2. (B) The EC1 interface with the A strand motif ‘SGWVW’ of the partner EC1 domain (C-atoms-green) contains two W residues. Only residues (black) that make favorable hydrophobic, van der Waals and hydrogen bond contact with the motif (red) are highlighted (H-bonds-dashed lines). (C) 3D structural model of celecoxib and (D) DMC with interactive residue side chains at the tryptophan W-binding pocket (F7, L24, S26, Y37, A75, A77, E87, S90, F92 and W4) are shown in stick rendering, with the carbon atoms of CDH sidechains colored white and the carbon atoms of the inhibitors colored green. The polypeptide backbones are rendered as ribbons. The red broken lines indicate potential intermolecular hydrogen bonds. Oxygen atoms are shown in red, flourine in pale green, nitrogen in blue, and sulfur in yellow. (E) Blocking CDH11 with celecoxib and (F) DMC significantly reduced the proliferation of CDH11 positive MDA-MB-231 as measured using MTS assay. (G) Native gel comparison of cadherin-11 EC1-2 in the absence (left) and presence (right) of celecoxib. Celecoxib was solubilized in DMSO and mixed with purified CDH11 EC1-2 in a 1:1 molar ratio. Note that celecoxib reduces the dimer fraction. (H) Recombinant modified CDH11 protein was immobilized on a Biacore® CM5 Surface by thiol coupling method. Wild type cadherin-11 was injected at various concentrations using Biacore T-200 instrument. Each concentration was injected twice, which showed good binding reproducibility. Colored lines represent real data-points and black lines represent curve fits. (I) 2D structure of the active compounds. (J)Sd-133 competed with CDH11 (ligand) in binding to immobilized CDH11 protein on the surface of the chip.