Fig. 3.
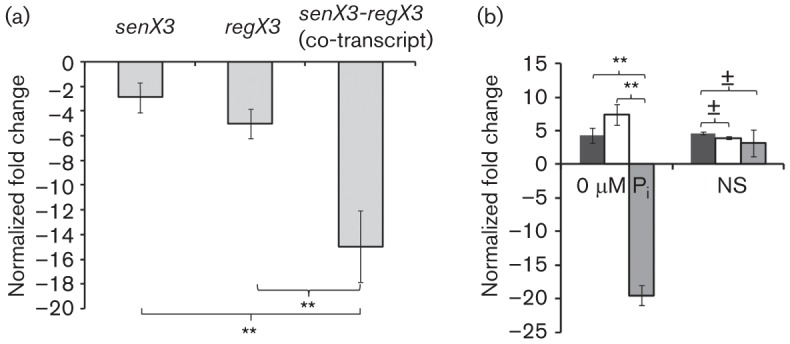
qRT-PCR reveals that bicistronic and monocistronic expression of Mtb senX3 and regX3 is condition-dependent. Total RNA was purified from wild-type CDC1551 grown in Middlebrook 7H9 broth to mid-exponential phase, and under nutrient starvation (NS) and phosphate depletion (0 µM Pi) conditions for 24 h. Abundance of transcripts of senX3 and regX3, and of senX3-regX3 co-transcripts was calculated relative to that of sigA. (a) Abundance of senX3-regX3 co-transcripts was significantly lower relative to that of senX3 (P<0.01) and regX3 (P<0.01) during growth in 7H9 broth. (b) Expression of senX3 (black bars) and regX3 (white bars) was upregulated, while that of the senX3-regX3 co-transcript (grey bars) was downregulated, during Mtb exposure to 0 µM Pi relative to 7H9 broth (P<0.01 for both comparisons). By contrast, expression of senX3, regX3 and the senX3-regX3 co-transcript was upregulated to a similar extent during Mtb exposure to NS (P>0.05 for both comparisons). The level of expression of each gene was normalized to that of the housekeeping gene sigA under each condition prior to comparison between individual stress conditions and 7H9 broth. Positive values in these graphs represent increased gene expression and negative values represent decreased expression under each stress condition relative to 7H9 broth. Samples were prepared in triplicate under each experiment condition. Error bars represent sd of the mean. ±, P>0.05; **P<0.01.
