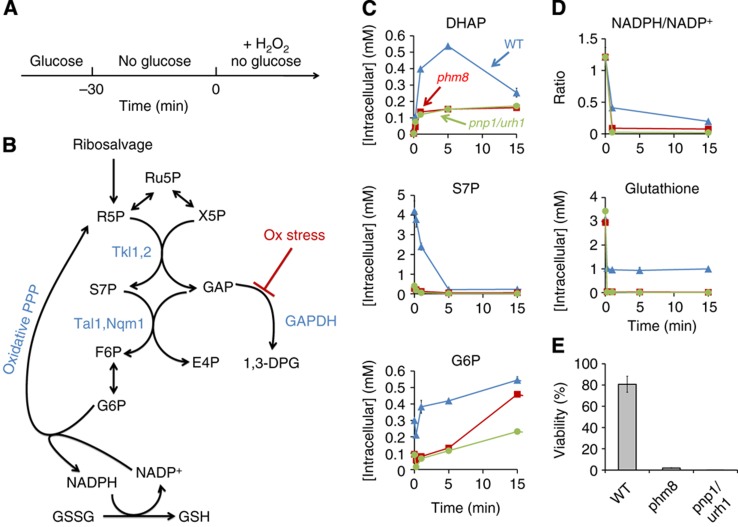
Figure 8.
The accumulation of S7P in carbon starvation facilitates survival of subsequent oxidative stress. (A) Experimental design. Yeast cells growing on glucose were switched to no carbon media for 30 min. Thereafter, 20 mM H2O2 was added and metabolome analyzed by LC-MS. (B) Diagram of proposed reactions and their regulation. S7P accumulates during carbon starvation due to low level of glyceraldehyde-3-phosphate (GAP). Oxidative stress blocks GAP consumption via glycolysis by inhibiting GAPDH. This provides sufficient GAP to react with S7P, producing F6P via transaldolase. F6P is further converted to G6P which enters the oxidative PPP. The resulting NADPH is utilized for regeneration of reduced glutathione. 1,3-DPG, 1,3-diphosphoglycerate; GSSG, glutathione disulfide; GSH, reduced glutathione. (C and D) DHAP, S7P, G6P, and glutathione levels and NADPH/NADP+ ratio in wild type, phm8 and pnp1/urh1 strains in the experiment shown in (A) The x axis represents minutes after oxidative stress, and the y axis represents absolute intracellular concentration or ratio of intracellular concentration (mean±range of N=2 biological replicates). (E) Viability of wild type, phm8 and pnp1/urh1 strains in oxidative stress. The y axis represents percentage of survived cells calculated by dividing the number of colonies formed after oxidative stress by the number of colonies formed under the same starvation condition but without H2O2 treatment (mean±range of N=2 biological replicates).