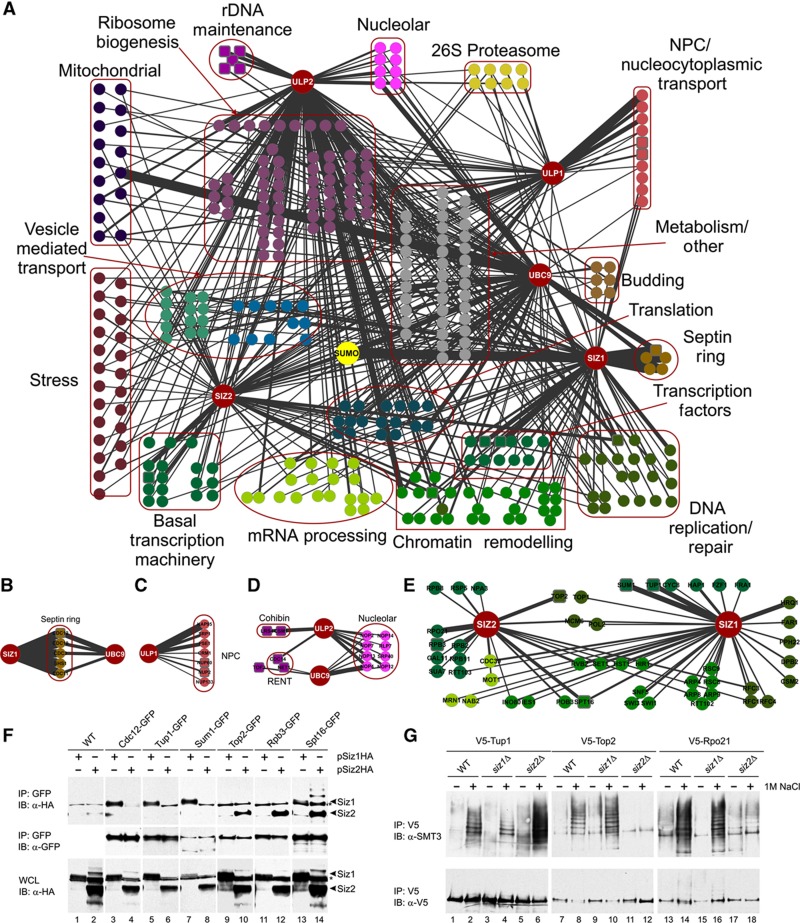
Figure 1.
(A) Functional organization of the budding yeast SUMO system. AP-MS was conducted to identify SUMO system component interactors. Large red nodes indicate proteins used as ‘baits’. Smaller nodes indicate interactors (‘prey’). Edge width is proportional to the average number of peptides identified for each prey protein. Square nodes indicate interactions confirmed using a second method. (B–E) Close-up of selected sub-networks. (B) Siz1 and Ubc9 localize to the septin ring during mitosis, and interact with septin proteins in our AP-MS. (C) Ulp1 localizes to the nuclear pore complex via interactions with several different karyopherins. (D) Ulp2 and Ubc9 interact with a number of nucleolar proteins, including components of the RENT and cohibin complexes. (E) The Siz1 and Siz2 interactomes are enriched for proteins involved in transcriptional control and chromatin remodeling. (F) Verification of Siz1 and Siz2 interactions via co-immunoprecipitation. GFP strains were transformed with Siz1- or Siz2-HA-ProtA mORF plasmids. GFP affinity purification was conducted, followed by immunoblotting using an antibody directed against HA (upper panel). An anti-GFP antibody was used to track the efficiency of each pulldown (middle panel), and whole-cell lysates (WCL) were evaluated with anti-HA to track the efficiency of protein induction (lower panel). A strain lacking GFP was used as negative control (WT). Siz1 and Siz2 migration are indicated by arrowheads. The asterisk indicates a non-specific band. (G) V5-tagged proteins identified as putative substrates in the AP-MS study were expressed in wt, siz1Δ and siz2Δ cells. Proteins of interest were subjected to V5 IP and western analysis, using antibodies directed against SUMO (top) or the V5 epitope (bottom).