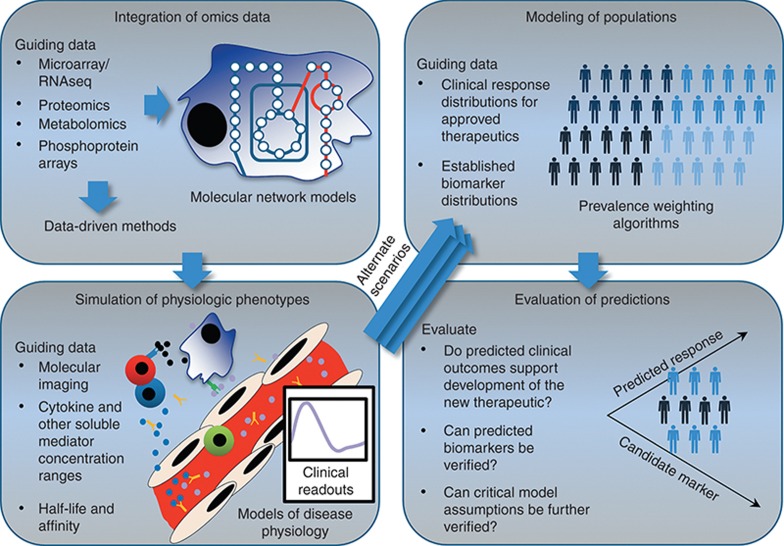
Figure 1.
Large data sets can be used to guide mechanistic models and ultimately develop quantitative predictions of clinical phenotypes at the population level. Omics data may be used to inform network models directly, or instead, to inform data-driven methods to assess cellular states. The resulting predictions for cellular function (cellular “exhaustion,” dysfunctional or inactive pathways, etc.) can be used to inform simulations of physiologic phenotypes that can be assessed and quantified in a clinical setting. Calibration or validation of the simulated physiologic phenotypes has been performed with additional data sets such as molecular imaging data and mediator concentrations. Given alternate physiologic parameters or alterations in the underlying network, alternate clinical outcomes are predicted. Disease outcome data can then be used to calculate the prevalence of the modeled patient phenotypes. The results are a prediction for the frequency of occurrence of underlying mechanisms in the disease pathophysiology, a rich simulated data set to mine for markers, and a validated disease-modeling platform that can be mechanistically altered to hypothesize and prioritize opportunities for therapeutic intervention. Portions adapted from refs. 3 and 9.