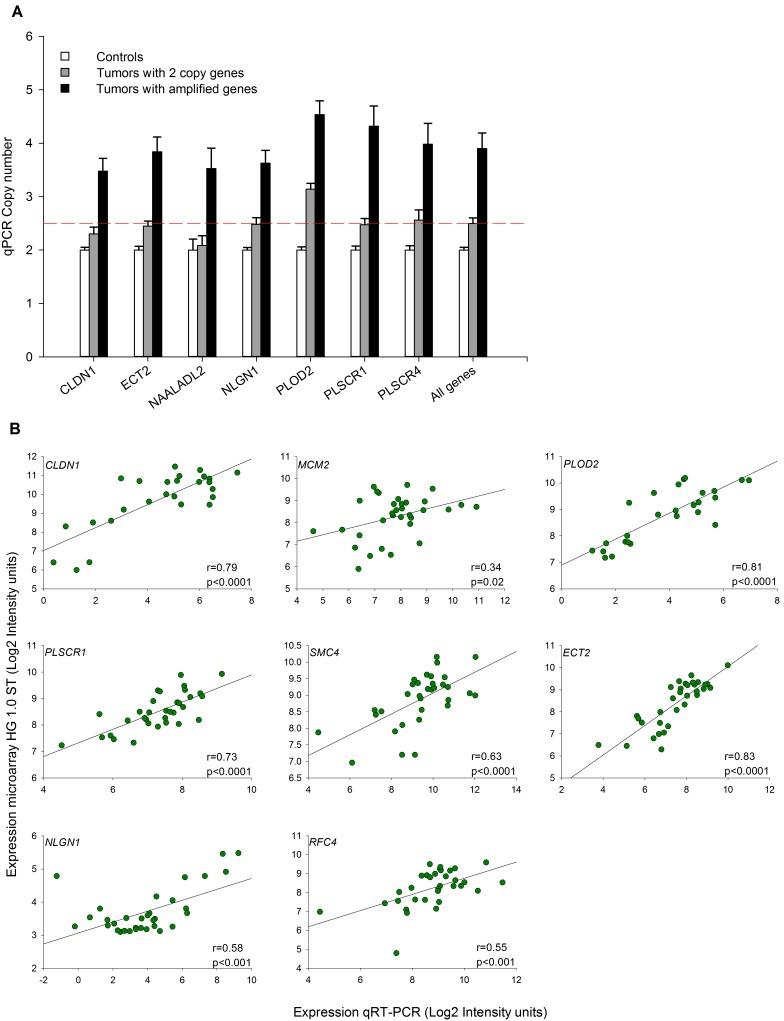
Figure 4. Validation of genes amplified and deregulated located in 3q by qPCR and qRT-PCR.
The top panel shows the average copy number of 7 genes (CLDN1, ECT2, NAALADL2, NLGN1, PLOD2, PLSCR1, and PLSCR4) located in 3q explored with qPCR in controls (lymphocytes) and tumors with 2 or 3–4 copies identified with the 500 K microarray. Whiskers of each bar represent the standard error of the mean. The dotted red line shows the value for 2.5 copies calculated with qPCR (see material and methods). The bottom panel shows the correlation of gene expression of 8 genes (MCM2, PLOD2, PLSCR1, SMC4, ECT2, NLGN1, RFC4, and CLDN1) located in 3q explored in 27 tumors and 6 controls with both the HG 1.0 ST microarray and qRT-PCR techniques. Log2 values of the normalized intensity signals obtained with the microarray (robust multichip average values) and qRT-PCR were plotted. Trend line (black line), correlation coefficient (r), and p value were calculated with Pearson’s correlation test.