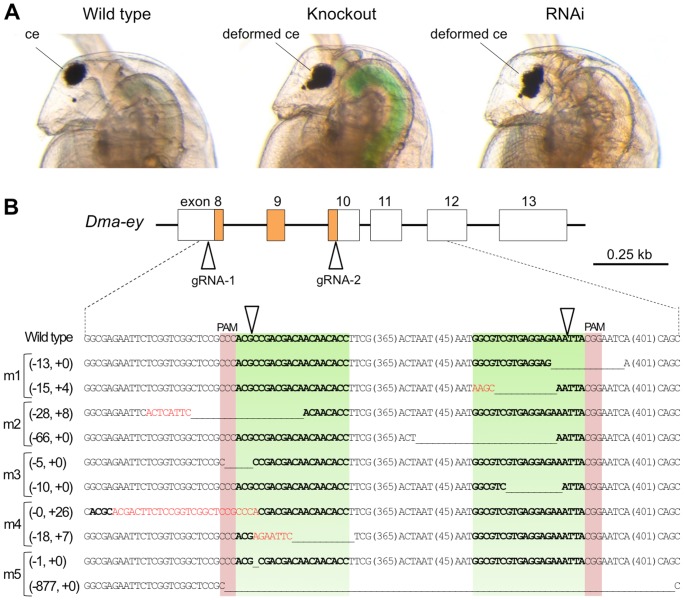
Figure 2. Knockout of Dma-eye gene.
(A) Typical phenotypes of Dma-ey deficient daphniids. The images to the left, center and right show the lateral head parts of the wild-type daphniid, Dma-ey knocked-out daphniid by the CRISPR/Cas system, and Dma-ey RNAi daphniid, respectively. The knocked-out individual is “mutant #5 (m5)” described in Figure 2B. Ventral side is left. ce: compound eye. (B) Genome sequences of established Dma-ey-knocked out mutant lines around gRNA-targeted sites. A part of the exon-intron structure of the Dma-ey gene including the homeobox colored in orange is shown above. In the alignment, the top line in the below alignment represents the wild-type Dma-ey sequence, and subsequent lines show sequences of five mutant alleles (see Table 1). The target sites for gRNAs are indicated in green, PAM in red, and cleavage site by triangles. The length (base pairs) of each in-del mutation is written in the left of each sequence (-, deletions; +, insertion). In each mutant sequence, deletion is indicated by underbars, insertions by red letters, sequences corresponding to the wild-type targeted sequences by bold letters, and the length (base pairs) of an abbreviated sequence within a parenthesis.