Figure 4.
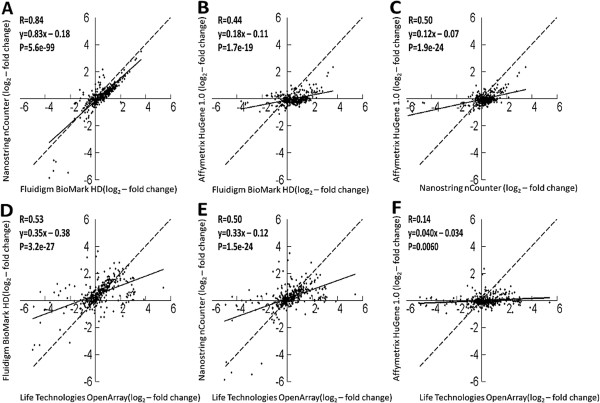
Fold change correlations between gene expression data quantified by the various platforms. Cross-platform comparisons were performed by evaluating the concordance of log2 fold changes by pair wise regression analysis of fold differences for chemical stimulations in comparison to vehicle controls. The average log2 fold changes for 28 transcripts present in all stimulations and detected on all three platforms were subjected to bivariate analysis. Subplots (A-F) illustrates the correlation between the following platforms: A) nCounter® and BioMark HD™, B) Affymetrix® and BioMark HD™, C) Affymetrix® and nCounter®, D) BioMark HD™ and OpenArray®, E) nCounter® and OpenArray®, F) Affymetrix® and OpenArray®. The solid line in each plot illustrates the linear regression fit while the dashed line represents the 1.0 slope of complete concordance. Pearson correlation coefficients (R) and equation for the linear fit (y) are indicated for each subplot. P-values for hypothesis of no correlation were calculated using Fisher’s transformation (P).
