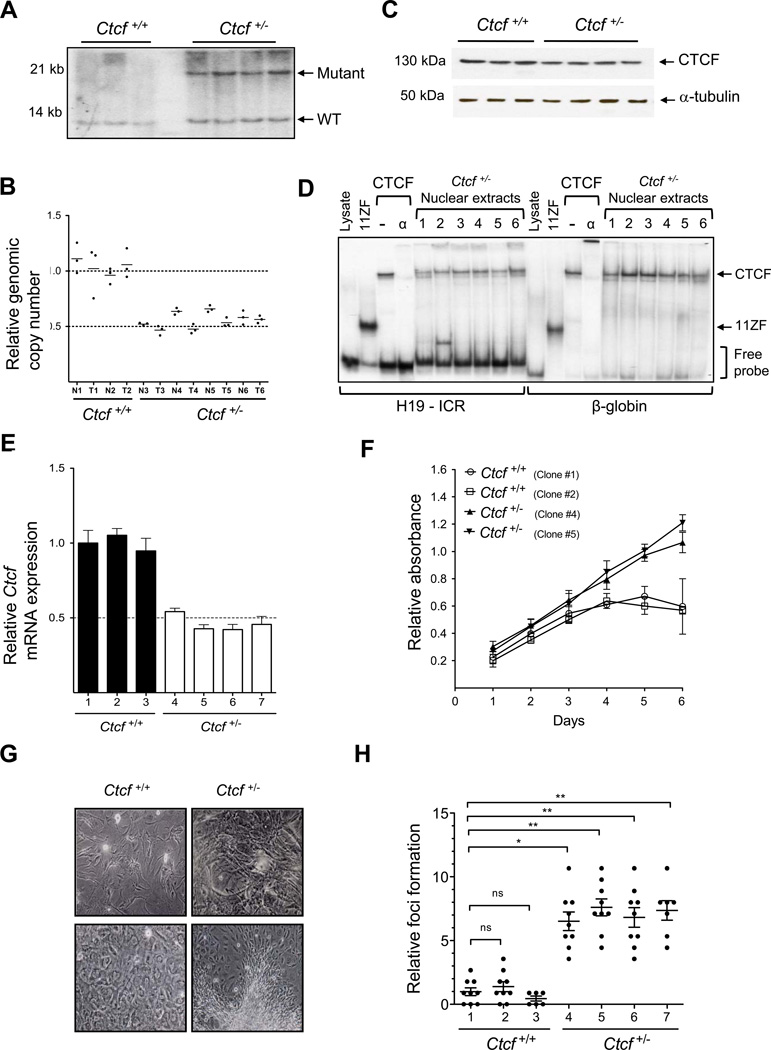
Figure 2. Ctcf is haploinsufficient for tumor suppression.
A, Southern blot analysis of lung tumors from Ctcf+/+ (lanes 1–3) and Ctcf+/− (lanes 4–7) mice. B, qPCR analysis of genomic DNA from normal lung (N) and lung tumors (T) from each genotype. C, Immunoblot analysis of CTCF protein in lung tumors from Ctcf+/+ (lanes 1–3) and Ctcf+/− (lanes 4–7) mice. α-tubulin served as loading control. D, Gel shift analysis of nuclear extracts from Ctcf+/− lung tumors show CTCF binding at both the H19/Igf2 ICR and β-globin insulator FII loci. Positions of protein-DNA complexes with 11ZF CTCF DNA binding domain (11ZF) or full length CTCF protein (CTCF) are indicated. α-CTCF antibody (α) was used to super-shift CTCF-DNA complexes. E, qRT-PCR analysis of Ctcf mRNA in Ctcf+/+ (n=3) and Ctcf+/− (n=4) MEFs, mean +/− s.e.m. F, Proliferation of Ctcf+/+ and Ctcf+/− MEFS. Assays performed in triplicate for two clones, each genotype; mean +/− s.d. G, Foci formation in MEFS cultured from Ctcf+/− compared to Ctcf+/+ mice. H, Increased foci formation in Ctcf+/− MEFs; mean +/− s.e.m., *P < 0.05, **P < 0.01.