Abstract
Rab GTPases are localized to various intracellular compartments and are known to play important regulatory roles in membrane trafficking. Here, we report the subcellular distribution and function of Rab14. By immunofluorescence and immunoelectron microscopy, both endogenous as well as overexpressed Rab14 were localized to biosynthetic (rough endoplasmic reticulum, Golgi, and trans-Golgi network) and endosomal compartments (early endosomal vacuoles and associated vesicles). Notably overexpression of Rab14Q70L shifted the distribution toward the early endosome associated vesicles, whereas the S25N and N124I mutants induced a shift toward the Golgi region. A similar, although less pronounced, redistribution of the transferrin receptor was also observed in cells overexpressing Rab14 mutants. Impairment of Rab14 function did not however affect transferrin uptake or recycling kinetics. Together, these findings suggest that Rab14 is involved in the biosynthetic/recycling pathway between the Golgi and endosomal compartments.
INTRODUCTION
Rab proteins constitute the largest subfamily of the low-molecular-weight GTPase superfamily and play key roles in the regulation of intracellular membrane trafficking. These proteins act as molecular switches that flip between a GDP-bound inactive state and a GTP-bound active form in which they recruit downstream effector proteins onto membranes. Rabs regulate diverse functions, including vesicle transport, by associating with cytoskeletal proteins; and vesicle targeting through interactions with tethering/docking factors (for reviews, see Zerial and McBride, 2001; Pfeffer, 2003). After Rab-mediated site-specific docking of an incoming vesicle to the target membrane, membrane fusion is achieved by the pairing of soluble N-ethylmaleimide-sensitive factor attachment protein receptors between lipid bilayers (Chen and Scheller, 2001).
Sequence analysis of the human genome indicates the presence of ∼60 different Rab genes (Bock et al., 2001). Similarly to soluble N-ethylmaleimide-sensitive factor attachment protein receptors, it is well established that different Rabs localize to distinct membrane-bound cellular compartments in mammalian cells, including the endoplasmic reticulum (ER)-Golgi region (Rab1a, Rab2, Rab6, Rab30, and Rab33b), early and recycling endosomes (Rab4, Rab5, Rab11, Rab15, Rab17, Rab18, Rab22, and Rab25), late endosomes/lysosomes (Rab7 and Rab9), and specialized organelles such as synaptic vesicles (Rab3a and 3c), secretory granules (Rab3D, Rab37), and melanosomes (Rab27) (reviewed in Zerial and McBride, 2001). Rabs associate with the membranes through a geranylgeranyl group linked to a cysteine residue(s) present in their carboxyl termini. Some Rabs are shown to be localized to more than one organelle: for example, Rab5 can be found at the plasma membrane, on clathrin-coated vesicles and endosomes (Gorvel et al., 1991; Bucci et al., 1992), and Rab11 has been detected on the trans-Golgi network (TGN) and recycling endosomes (Urbé et al., 1993; Ullrich et al., 1996).
Endosomal Rabs (Rabs 4, 5, 7, 9, and 11) are the best characterized with respect to their localization and functional interaction with effector proteins. Rab5 plays an important role in the clathrin-mediated endocytosis of molecules (receptors and ligands) from the cell surface and in homotypic endosome-endosome fusion (Gorvel et al., 1991; Bucci et al., 1992). Rab4 and Rab11 regulate the recycling of receptors from early endosomes to the cell surface by distinct pathways: Rab4 is suggested to function at the level of the early sorting endosomes (van der Sluijs et al., 1992; de Wit et al., 2001) and Rab11 in the trafficking of cargo through perinuclear recycling endosomes (Ullrich et al., 1996; Ren et al., 1998). Several Rab effectors have been identified, including Rabaptin-4 and -5, Rabip4/RUFY1 for Rab4 (Vitale et al., 1998; Nagelkerken et al., 2000; Cormont et al., 2001; Yang et al., 2002); Rabaptin-5, Rabex, Rabinosyn, EEA1, and phosphatidylinositol-3-OH kinases for Rab5 (Horiuchi et al., 1997; Gournier et al., 1998; Christoforidis et al., 1999a,b; Nielsen et al., 2000); and FIP family members for Rab11 (Prekeris et al., 2000, 2001; Hales et al., 2001; Lindsay and McCaffrey, 2002). Rab7 plays a role in transport from early-to-late endosomes and late endosomes-to-lysosomes by interacting with its effector protein RILP (Cantalupo et al., 2001). Rab9 is thought to regulate the late endosome-to-TGN transport step through its interaction with TIP47 (Carroll et al., 2001).
Even though the localization of several Rab proteins and their function in vesicular transport are well documented, the role of various other Rab proteins has yet to be elucidated. The cDNA cloning of Rab14 from rat brain was reported by our laboratory previously (Elferink et al., 1992), but its function in membrane trafficking is currently unknown. Amino acid sequence alignments reveal that Rab14 is a close homolog of Rab2 and Rab4 (Bock et al., 2001). It is well established that these two GTPases function in the Golgi and endosomal compartments, respectively (for reviews, see Zerial and McBride, 2001). We therefore hypothesized that Rab14 functions in one or both of these compartments. To test this hypothesis, we have generated a Rab14-specific polyclonal antibody and have used it to show that endogenous Rab14 is localized to both the biosynthetic (ER, Golgi, and TGN) as well as endosomal compartments by light and electron microscopy immunolocalization studies. Overexpression of green fluorescent protein (GFP)-tagged Rab14 either as wild-type or in the Q70L mutant shifts the Rab14 distribution to the endosomal compartments, whereas overexpression of the S25N and N124I causes a shift toward the Golgi region. Notably, this localization of the distinct Rab14 mutants is mimicked by the distribution of transferrin receptor (TfR), although with more moderate effects. Our results suggest that Rab14 may play a role in biosynthetic as well as recycling pathways between the Golgi and endosomal compartments.
MATERIALS AND METHODS
Plasmids
The open reading frame of rat Rab14 (GenBank accession no. M83680) was amplified by polymerase chain reaction (PCR) from rat brain cDNA library (BD Biosciences Clontech, Palo Alto, CA) and subcloned into a pGEX-KG vector. Similarly Rab1a (human), Rab2 (rat), Rab3a (rat), Rab4a (human), Rab10 (mouse), Rab15 (rat), Rab16/3d (rat), and Rab17 (mouse) were subcloned into the pGEX-KG vector. The Rab14 cDNA was also subcloned into pEGFP-c1 vector (BD Biosciences Clontech). Rab14S25N, Rab14Q70L, and Rab14N124I mutants were generated by PCR mutagenesis by using wild-type pEGFP-Rab14 as a template. Rab4a and Rab11a cDNAs (human) were subcloned into a pCMV-3c vector (Stratagene, La Jolla, CA) with an N-terminal myc tag.
Production of Glutathione S-Transferase (GST)-Rab Proteins
pGEX-Rab construct was transformed into BL21(DE3) RIL strain (Stratagene), cells were grown at 37°C and induced with 0.5 mM isopropyl thio-β-d-galactopyranoside for 4 h. Cells were harvested, resuspended in phosphate-buffered saline (PBS) containing 5 mM 2-mercaptoethanol and 1% Triton X-100 and lysed using a microfluidizer. The lysate was centrifuged for 30 min at 28,000 × g. The supernatant was loaded onto a glutathione-Sepharose (Sigma-Aldrich, St. Louis, MO) column at 4°C, and the column was washed with PBS containing 5 mM 2-mercaptoethanol and 0.1% Triton X-100. The GST-Rab was eluted with 10 mM glutathione and dialyzed against PBS. In the case of GST-Rab14, the Rab was cleaved from GST beads by thrombin (Calbiochem, La Jolla, CA) for the production of polyclonal antibodies.
Antibodies
Rabbit polyclonal antibodies were raised against thrombin-cleaved Rab14 protein (Josman LLC, Napa, CA). GST-Rab4a, GST-Rab14, and GST-Rab15 were coupled to cyanogen bromide-activated Sepharose 4B beads. Rabbit serum against Rab14 was first loaded onto a GST-Rab15 column; the filtrate was then loaded onto a GST-Rab14-Sepharose column. Bound antibodies were eluted with 0.1 M glycine (pH 2.3), neutralized with Tris, and dialyzed against PBS. The Rab14 affinity-purified antibodies were finally incubated with GST-Rab4a-Sepharose beads for 1 h at room temperature to remove any nonspecific antibodies, and the supernatant was stored in the presence of 50% glycerol at –80°C.
The mouse monoclonal antibodies used in this study are anti-EEA1, anti-GM130, anti-clathrin, and anti-TGN38 (BD Transduction Laboratories, Lexington, KY); anti-human TfR (Zymed Laboratories, South San Francisco, CA); anti-myc (9E10) (Santa Cruz Biotechnology, Santa Cruz, CA), and anti-human LAMP-1 (H4B3) and LAMP-2 (H4B4) (Developmental Studies Hybridoma Bank, University of Iowa, Iowa City, IA). Rabbit anti-syntaxin13 was described previously (Prekeris et al., 1998). The rabbit anti-lgp120/LAMP-1 antibody M3 was a gift from I. Mellman. Rabbit anti-sec23 was obtained from Affinity Bioreagents (Golden, CO). Rabbit anti-GFP and secondary antibodies (Alexa-488- or Alexa-594–conjugated anti-rabbit or anti-mouse) were purchased from Molecular Probes (Eugene, OR), and unconjugated rabbit anti-mouse antibodies from Dako (Glostrup, Denmark).
Tissue Expression Analysis for Rab14 by PCR and Western Blot
PCR Analysis. Human multiple tissue cDNA panels were purchased from BD Biosciences Clontech and used as template DNA for PCR analysis. Two primers were designed, a forward primer (5′-ATG GCA ACT GCA CCA TAC AAC TAC TCT TAC-3′) for the Rab14 open reading frame (amino acids 1–10) and a reverse primer (5′-TGA AAG GTC AAA TGA GGG GCC ACA GCA AAG-3′) for the 3′ untranslated region. The design of the reverse primer in the 3′ untranslated region significantly reduces the risk of amplifying related members of the same gene family to allow unambiguous analysis of gene expression. The PCR analysis was carried out using the TITANIUM TaqPCR kit (BD Biosciences Clontech) according to the manufacturer's instructions with glyceraldehyde-3-phosphate dehydrogenase as a positive control for constitutive expression. After 25 cycles, samples were analyzed on agarose gel.
Western Blot Analysis. Rat organs were homogenized in 3 ml of HB buffer (20 mM HEPES, pH 7.2, 10 mM sucrose, 10 mM KCl, 2 mM EDTA, 2 mM EGTA, 6 mM MgCl2, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride, 2 μg/ml leupeptin, 4 μg/ml aprotinin, 0.7 μg/ml pepstatin A) per gram in a Teflon-glass homogenizer. Postnuclear supernatant (PNS) was produced by spinning at 2000 × g for 10 min. For the Western blot in Figure 2B, equal total PNS concentrations of different tissues were loaded on 4–20% SDS-PAGE gel. The Western blot was incubated with primary anti-Rab14 antibody, followed by incubation and detection with a standard horseradish peroxidase-labeled secondary antibody detection system (Pierce Chemical, Rockford, IL).
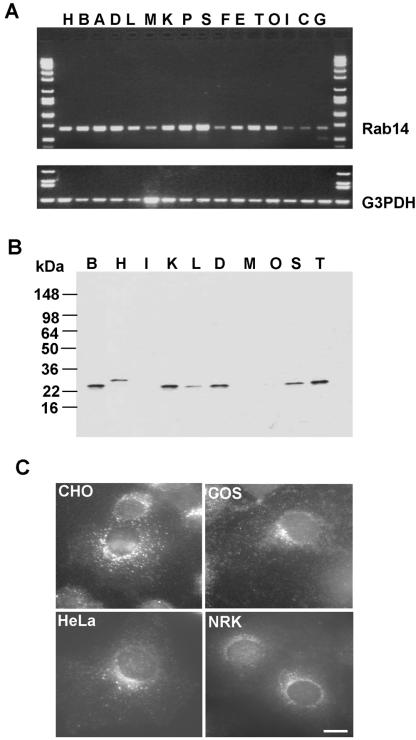
Figure 2.
Rab14 GTPase is ubiquitously expressed in all tissues and cell types. (A) Probing expression of Rab14 gene at mRNA level by PCR analysis by using BD Biosciences Clontech human multiple tissue cDNA panels as template (see MATERIALS AND METHODS for more details). Top, corresponds to the Rab14 PCR product. Bottom, corresponds to the glyceraldehyde-3-phosphate dehydrogenase (G3PDH) PCR product. (B) Various rat tissue protein extracts (PNSs) were analyzed by Western blotting by using Rab14 polyclonal antibodies. A single 24-kDa protein band was detected in most of the samples with highest expression levels in brain, kidney, lung, spleen, and thymus. Lanes represented in A and B: A, placenta; B, brain; C, colon; D, lung; E, prostate; F, thymus; G, leukocyte; H, heart; I, small intestine; K, kidney; L, liver; M, muscle; O, ovary; P, pancreas; S, spleen; T, testis. (C) Rab14 localization in different cell lines. Cells were fixed with 3% PFA, permeabilized with saponin, and stained using affinity-purified Rab14 antibody followed by incubation with Alexa-488–labeled anti-rabbit IgG antibodies. Bar, 10 μm.
Cell Culture and Transient Transfection
Normal rat kidney (NRK) and HeLa cells were grown in DMEM supplemented with 10% fetal bovine serum, 2 mM l-glutamine, 100 U/ml penicillin G, 100 μg/ml streptomycin in a 5% CO2 incubator at 37°C. The cells were transfected with LipofectAMINE 2000 (Invitrogen, Carlsbad, CA) for 18–24 h and then processed for immunofluorescence microscopy, immunoelectron microscopy, or for transferrin uptake and recycling assays.
Immunofluorescence Microscopy
Cells were grown on eight-chamber slides (Lab-Tek, Naperville, IL) and fixed with 3% paraformaldehyde (PFA) in PBS for 15 min, followed by two 10-min incubations in 0.1 M glycine/PBS. The cells were permeabilized with PBS containing 0.4% saponin, 2% fetal bovine serum, and 1% bovine serum albumin for 30 min and then incubated with primary antibodies for 1 h at room temperature. After extensive washing, cells were incubated with Alexa-conjugated secondary antibodies for 30 min, washed and mounted with Vectashield (Vector Laboratories, Burlingame, CA). Cells were imaged with an Axiovert 200 fluorescence microscope, fitted with a charge-coupled device camera (both from Carl Zeiss, Thornwood, NY).
Immunoelectron Microscopy
NRK cells, either nontransfected or transfected with the different Rab14-GFP constructs, were fixed for 2.5 h at room temperature in 2% PFA and 0.2% glutaraldehyde in 0.1 M phosphate buffer at pH 7.4. After storage in 2% PFA, the cells were rinsed with PBS, scraped, and embedded in 12% gelatin. Small blocks of embedded material were infiltrated overnight with 2.3 M sucrose at 4°C, mounted on aluminum pins and frozen in liquid nitrogen. Ultrathin sections were cut at –120°C, and picked up with a solution of 1% methylcellulose (Sigma-Aldrich) and 1.2 M sucrose in phosphate buffer. After washing with PBS containing 0.02 M glycine, the sections were single or double labeled with antibodies and protein A-gold as described previously (Slot et al., 1991). Control labelings with GFP antibodies on nontransfected cells and with secondary antibodies alone were negative.
Quantification of the Rab14 distribution was done by counting gold particles on sections of NRK cells labeled either with anti-Rab14 (for endogenous Rab14) or with anti-GFP (for overexpressed GFP-Rab14 forms). Gold particles within 25 nm of a membrane were scored as belonging to that particular organelle. Vesicles and tubules within 200 nm from the Golgi stack or an early endosome (EE) vacuole were designated as Golgi- or EE-associated vesicles, respectively. For each Rab14 form, three counting sessions on randomly sampled section fields were performed, each yielding at least 300 gold particles bound to membranes. The quantity of gold particles associated with each organelle type was expressed as the percentage of the total number of membrane-bound gold particles for each session. Average percentages (±SEM) of the three sessions are represented.
To quantify the TfR distribution in cells transfected with GFP-Rab14Q70L or GFP-Rab14S25N, cell profiles with Rab14 overexpression were randomly sampled on sections of NRK cells double labeled for TfR and GFP. Only cell profiles displaying a nucleus, a Golgi stack, and an EE vacuole, and with at least 100 Rab14-representing gold particles were analyzed. Quantification of TfR labeling was performed as for Rab14. Respective percentages for each cell profile were averaged for each Rab14 form and compared by means of Student's t test.
Transferrin Uptake and Recycling
Transiently transfected HeLa cells were plated on collagen-coated glass coverslips, washed with PBS, and incubated for 1 h at 4°C in serum-free, HEPES-buffered DMEM with 20 μg/ml Alexa 594-conjugated transferrin (Molecular Probes) followed by incubation at 37°C at various time intervals (5, 20, and 60 min). Cells were fixed with 3% PFA and imaged. For recycling assays, after 30 min of Alexa 594-conjugated transferrin uptake, cells were extensively washed with serum free DMEM and incubated with 200 μg/ml unlabeled transferrin at 37°C for various time intervals, fixed, and imaged. For fluorescence-activated cell sorting analysis, cells grown on 10-cm Petri dishes were used for transient transfections. After 16 h posttransfection, cells were trypsinized and incubated with Alexa 647-conjugated transferrin as described above for uptake as well as recycling assays. Cells were fixed with 2% PFA, analyzed, and sorted for transfected cells using GFP fluorescence, and the transferrin present in the GFP-containing cells was quantitated using Alexa 647 fluorescence.
RESULTS
Sequence Comparison and Tissue Expression Profile of the Rab14 Gene
The predicted Rab14 amino acid sequence consists of 215 amino acids and was originally identified from a rat brain cDNA library (Elferink et al., 1992). It is essentially identical to mouse and human Rab14 orthologues except in the fourth amino acid position, where Thr in the rat sequence is replaced by an Ala residue. Rab4 (endosomes) and Rab2 (Golgi) are the closest homologues of Rab14 with 58%/67% and 57%/68% identity/similarity, respectively.
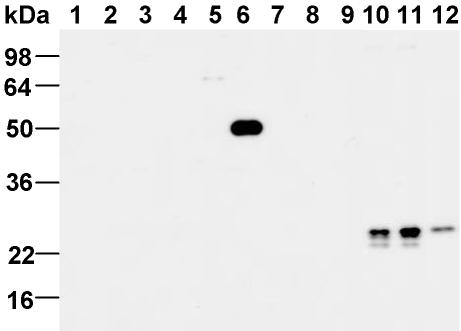
To study the subcellular localization of the Rab14 protein, we generated rabbit polyclonal antibodies against recombinant full-length Rab14 that was produced in Escherichia coli (see MATERIALS AND METHODS for details). As shown in Figure 1, the affinity-purified antibody specifically recognized the GST-Rab14 protein, but none of the other Rabs tested, including Rab1, Rab2, Rab3, Rab4, Rab10, Rab15, and Rab17. The antibody detected a single 24-kDa protein from rat brain cytosol, postnuclear supernatant (cytosol + membrane), and NRK total cell extract, in agreement with its the predicted molecular weight of Rab14. The detection of endogenous Rab14 protein from cell extracts is abolished by preincubation of the antibody with recombinant Rab14 protein (our unpublished data), indicating that the purified antibody is specific to Rab14.
Figure 1.
Specificity of Rab14 antibodies. GST-Rabs (100 ng) were analyzed on SDS-PAGE, transferred onto a nitrocellulose membrane, and probed with a Rab14 polyclonal antibody. Lanes 1–9 correspond to Rab1a, Rab2, Rab3a, Rab4a, Rab10, Rab14, Rab15, Rab16/3d, and Rab17, respectively. Lane 10, rat brain cytosol (60 μg), lane 11, rat brain PNS (60 μg), and lane 12, NRK total cell extract (30 μg). As expected, no detectable signal was observed with any other GST-Rabs other than Rab14.
The tissue expression pattern of Rab14 at the mRNA level was studied by PCR analysis as described under MATERIALS AND METHODS. As seen in Figure 2A, the expected 680-base pair Rab14 PCR product was obtained in all tissues, with relatively higher yields in brain, heart, kidney, placenta, lung, pancreas, spleen, and testis samples. However, lower levels of PCR products were obtained from muscle, thymus, intestine, colon, and leukocyte samples. To further analyze the tissue distribution of Rab14 protein, the affinitypurified polyclonal antibody was used to probe the Western blot of various rat tissue extracts. In agreement with the PCR analysis, the antibody detected a single 24-kDa protein band in most tissue samples, with relatively high protein levels in brain, kidney, lung, spleen, and testis (Figure 2B). A slightly larger protein (∼28 kDa) was observed in heart tissue compared with all other tissue samples. The antibody also stained all the cell lines tested (Chinese hamster ovary, COS7, HeLa, and NRK), and the endogenous Rab14 was detected in small puncta throughout the cell as well as in perinuclear structures (Figure 2C). The ubiquitous expression of Rab14 suggests a fundamental role for this protein in a membrane trafficking event common to all cell types.
Subcellular Localization of Endogenous Rab14
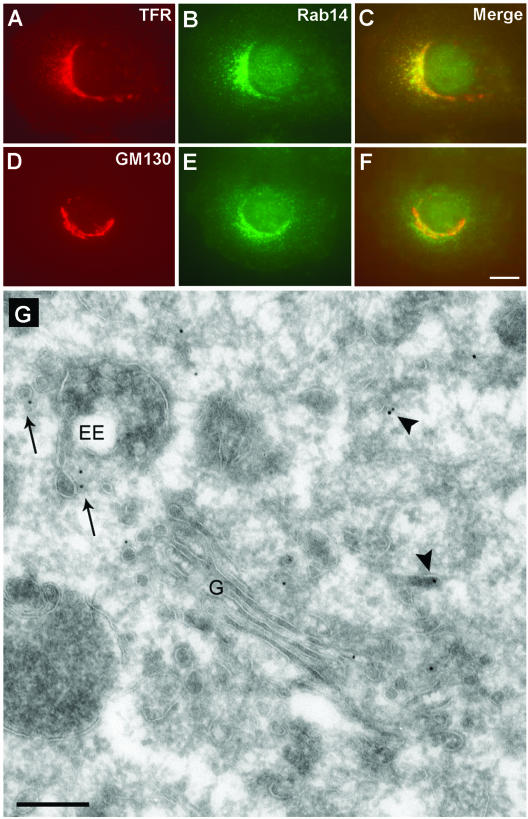
An important step in understanding the function of a membrane trafficking protein is to identify its subcellular localization. Toward this objective, we coimmunostained endogenous Rab14 in NRK cells with well-characterized markers of Golgi, endosomal, and lysosomal compartments by immunofluorescence microscopy. Rab14 staining partially overlapped with TfR, a marker for EEs and recycling endosomes (REs), and to some extent with the cis-Golgi and TGN markers GM130 and TGN38 (Figure 3; our unpublished data). There is no overlap of Rab14 in HeLa cells with the late endosomal/lysosomal markers LAMP-1 and LAMP-2 or in NRK cells with LysoTracker Red, indicating that Rab14 is not associated with the late endosomal/lysosomal compartments (our unpublished data).
Figure 3.
Subcellular localization of endogenous Rab14 protein. NRK cells were fixed with 3% PFA, permeabilized with saponin, and costained using rabbit anti-Rab14 antibody (B and E) and mouse monoclonal antibodies against TfR (A), GM130 (D), followed by incubation with Alexa-488–labeled anti-rabbit and Alexa-594–labeled anti-mouse IgG antibodies. (C and F) Merged images of A and B and D and E, respectively. Bar, 10 μm. (G) Ultrathin cryosection of an NRK cell, immunogold-labeled for Rab14 (10-nm gold). Rab14 is present on the Golgi stack (G) and associated vesicles, in vesicles (arrows) nearby early EEs and in more dispersed tubulo-vesicular structures (arrowheads). Bar, 200 nm.
To examine the subcellular distribution of Rab14 in greater detail, we prepared NRK cells for immunoelectron microscopy. Immunogold staining of ultrathin cryosections yielded a low but significant labeling for endogenous Rab14 (Figure 3G). Quantitation of the labeling pattern corroborated the finding that Rab14 is associated both with membranes of the biosynthetic as well as the endocytic pathways (Table 1). In agreement with the immunofluorescence data, 19% of Rab14 label was found on the Golgi stack and associated vesicles. Surprisingly, the endoplasmic reticulum also showed significant Rab14 staining. A minor fraction of label was found on the limiting membrane of endosomal vacuoles, and on vesicles/tubules in proximity to the vacuole. The major fraction (≥50%) of membrane-bound endogenous Rab14 in NRK cells, however, was localized to small (40–60 nm) tubulo-vesicular structures that were found dispersed in the cytoplasm. These are likely to shuttle between the Golgi and EEs. Remarkably, a small but significant portion of Rab14 was also found on the plasma membrane.
Table 1.
Relative distribution of endogenous and overexpressed wild-type and mutant Rab14
| Endogenous Rab14 | Rab14wt | Rab14Q70L | Rab14S25N | |
|---|---|---|---|---|
| RER+ nuclear envelope | 10.7 ± 1.3 | 7.8 ± 1.8 | 5.1 ± 1.1 | 15.3 ± 3.9 |
| Golgi stack | 12.9 ± 2.4 | 17.7 ± 2.8 | 6.0 ± 1.7 | 32.1 ± 2.5 |
| Vesicle/tubules Golgi region | 5.8 ± 1.5 | 6.2 ± 1.7 | 4.4 ± 0.9 | 9.2 ± 0.8 |
| Early endosomal vacuoles | 5.8 ± 1.4 | 8.1 ± 0.8 | 14.7 ± 3.0 | 4.6 ± 0.6 |
| Vesicles/tubules near EE | 5.3 ± 1.2 | 11.5 ± 1.6 | 17.3 ± 2.8 | 2.9 ± 0.5 |
| Late endosomes/lysosomes | 1.8 ± 0.5 | 1.8 ± 0.6 | 2.5 ± 1.1 | 2.8 ± 1.4 |
| Cytoplasmic vesicles/tubules | 44.4 ± 1.5 | 45.2 ± 1.4 | 46.8 ± 3.4 | 28.3 ± 3.5 |
| Plasma membrane | 13.3 ± 1.6 | 1.7 ± 0.2 | 3.3 ± 0.8 | 4.8 ± 0.9 |
| Total membrane-associated gold | 1095 | 1216 | 1048 | 1198 |
Numbers represent the percentage of membrane-bound Rab14 that is associated with a given compartment and represent averages of three counting sessions ± SEM. The total membrane-associated gold is the sum of all membrane-associated gold particles from the three counting sessions.
Localization of GFP-Rab14 Fusion Proteins in Live NRK Cells
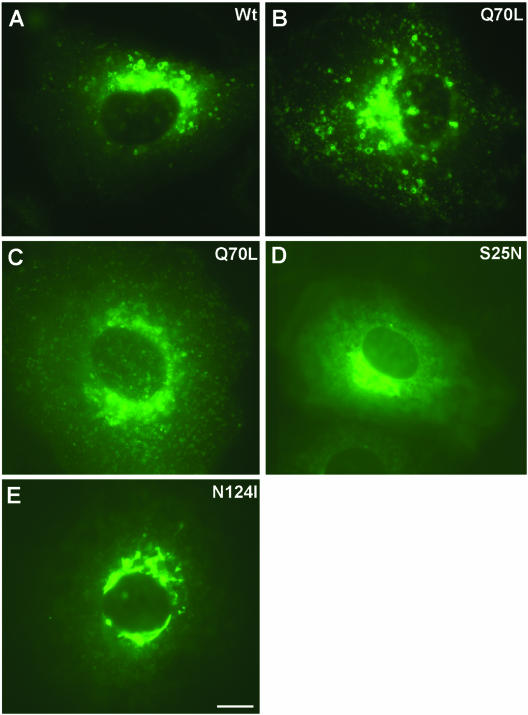
Like all other small GTPases, Rabs are generally considered constitutively active when bound to GTP and inactive when in a GDP-bound state. Mutations within the three conserved motifs, phosphate/Mg2+ (PM) interacting loop 1 (Ser/Thr to Asn), G2 (Asn to Ile), and PM3 (Gln to Leu) perturb the cycle of nucleotide binding and hydrolysis, thereby interfering with specific membrane transport steps in a dominant manner. For some Rab proteins, the biochemical properties with respect to nucleotide binding and hydrolysis of these mutations are well established: the PM1 (Ser/Thr to Asn) mutant retains a GDP-bound conformation and the G2 (Asn to Ile) mutant is in a nucleotide free form, whereas the PM3 mutant (Gln to Leu) is in a GTP-bound conformation (Vitelli et al., 1997; Zuk and Elferink, 2000; Wilcke et al., 2000). To more closely examine the function of Rab14, we have generated site-specific mutants of corresponding residues in Rab14 (Rab14-S25N, -N124I, and -Q70L) and transiently overexpressed each in NRK cells as GFP fusion proteins. GFP-Rab14wt and GFP-Rab14Q70L proteins showed similar fluorescence imaging patterns, with fluorescent spots (representing vesicles, or clusters of vesicles) present throughout the cell but more concentrated in the perinuclear region (Figure 4, A and B). The size of these vesicles/fluorescent spots was dependent on Rab14 expression level: the larger vesicles, showing up as fluorescent rings, were observed only in cells with high levels of expression (compare Figure 4B with C). Similar results were observed in HeLa, Chinese hamster ovary, and COS7 cell lines (our unpublished data). In contrast, the GFP-Rab14S25N and GFP-Rab14N124I associated with Golgi-like perinuclear structures (Figure 4, D and E).
Figure 4.
Detection of overexpressed GFP-Rab14 fusion proteins in live NRK cells by fluorescence microscopy. Live NRK cells were imaged using GFP fluorescence after 24 h of transfection with various GFP-Rab14 constructs. (A) GFP-Rab14wt. (B and C) GFP-Rab14Q70L. (D) GFP-Rab14S25N. (E) GFP-Rab14N124I. Large vesicles were formed in GFP-Rab14 wt and Q70L cells. The GFP signal was more diffuse and localized to perinuclear Golgi-like structures in GFP-Rab14S25N and N124I cells. Note that the cell shown in B expresses a moderate-to-high level of GFP, whereas that in C expresses low-to-moderate level. Bar, 10 μm.
Rab14wt and -Q70L Overexpression Induces the Appearance of Rab14-positive Membranes near EEs
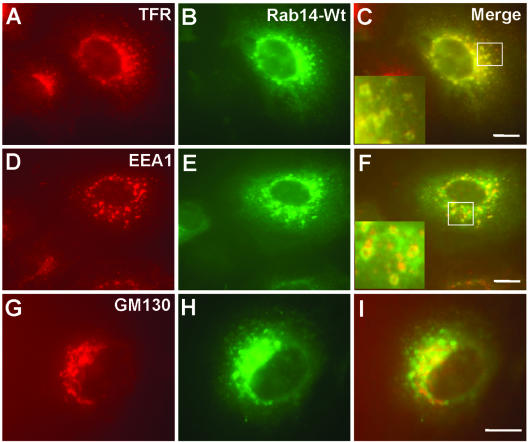
To determine the identity of vesicles associated with overexpressed Rab14wt or Q70L mutant, transfected cells were analyzed both by immunofluorescence and immunoelectron microscopy by using well-characterized marker protein antibodies. The applied markers were EEA1 (for EEs), TfR (for EEs and REs), GM130 and TGN38 (for the Golgi apparatus and TGN), LAMP-1 and LAMP-2 (for late endosomes/lysosomes), and syntaxin13 (for REs). As evidenced in Figure 5, the larger vesicles/spots stained for EEA1 and TfR, defining these as EEs. Overlap of the GFP signal with TfR staining was observed in the perinuclear region, suggesting that GFP-Rab14 was also localized to the RE region of the endosomal compartment. As with endogenous Rab14, there was also partial overlap of GFP-Rab14wt with GM130. There was no costaining of LAMP-1 and LAMP-2 markers with GFP-Rab14wt signal, although there was a little overlap in the case of the GFP-Rab14Q70L mutant (our unpublished data).
Figure 5.
GFP-Rab14wt fusion protein is primarily localized to TfR-positive compartments. After PFA fixation and saponin permeabilization, GFP-Rab14 wt cells were stained with mouse monoclonal antibodies, TfR (A), EEA1 (D), and GM130 (G), followed by incubation with Alexa-594–labeled anti-mouse IgG antibody. Green corresponds to the GFP signal (B, E, and H). (C, F, and I) Merged images. Note that GFP-Rab14 colocalizes with TfR much better than with EEA1. Partial colocalization of GFP-Rab14 is seen with the Golgi marker GM130. Bars, 10 μm.
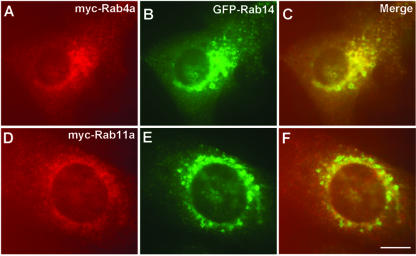
The endosomal compartment can be subdivided into Rab4/Rab5-positive and Rab4/Rab11 subdomains. Internalized transferrin has been shown to move from Rab5-positive to Rab5/Rab4 endosomes and then through Rab4/Rab11 containing recycling endosomes back to the cell surface (Sönnichsen et al., 2000). To determine with which endosomal domain Rab14 associates, we cotransfected GFP-Rab14 wt with myc-tagged constructs of either Rab4 or Rab11 and studied their colocalization by immunostaining with an anti-myc antibody. As seen in Figure 6, the overlap between Rab14-positive and Rab4-positive endosomes was stronger than with Rab11-positive endosomes.
Figure 6.
Rab4 colocalizes with Rab14-positive organelles. NRK cells were cotransfected with GFP-Rab14wt and myc-Rab4a or myc-Rab11a. Rab4 and Rab11 are localized by staining with mouse anti-myc antibody (9E10) followed by incubation with Alexa-594–labeled anti-mouse IgG antibody. Green panels show the Rab14 localization (B and E), and red panels show the localization of Rab4 (A) and Rab11 (D). Note that Rab14 colocalized with Rab4 on same vesicles/fluorescence spots, but not with Rab11. Bars, 10 μm.
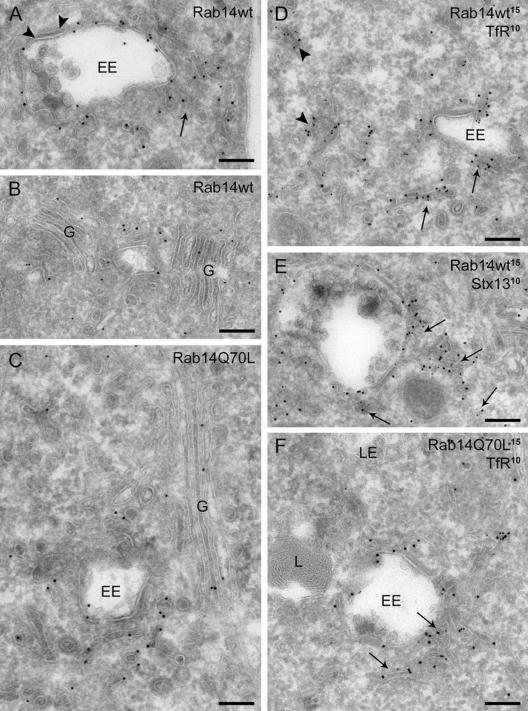
Labeling of ultrathin cryosections for immunoelectron microscopy with anti-GFP revealed that the membrane-bound fraction of the GFP-Rab14wt construct has the same localization pattern as endogenous Rab14, albeit with significant lower staining of the plasma membrane (Table 1). GFP-Rab14wt was mainly found on narrow (diameter 40–60 nm) tubules and vesicles dispersed in the cell or close to EEs. Particularly in cells with high expression levels, the number of tubulo-vesicular structures associated with EEs was clearly enhanced in comparison with nontransfected cells (Figure 7A). This zone of accumulated Rab14wt-positive vesicles around the EEs probably accounts for the ring-like appearance of vesicles in the fluorescence images. The majority of Rab14 label on tubular profiles was along noncoated membrane domains. The limiting membrane of EEs was only sparsely labeled, with virtually no label on the bilayered coat covering part of the EE perimeter. Additional label was present on the Golgi stack and adjacent vesicles and tubules on both sides of the stack (Figure 7B). Double labeling with antibodies against GFP and sec23 or clathrin as markers for the vesiculotubular clusters (VTCs) or TGN, respectively, showed that Rab14 is present both in the VTCs and the TGN (our unpublished data).
Figure 7.
Overexpression of Rab14wt and Q70L causes accumulation of Rab14-positive tubules and vesicles near EEs; and Rab14Q70L shifts from the Golgi toward the EE region. (A) Overexpressed GFP-Rab14wt is abundantly present on tubules clustered at the periphery of EEs (arrow). Some label is found at the limiting membrane of EEs, but not on the bilayered coated domain (arrowheads). (B) Additional label for GFP-Rab14wt is found on the Golgi cisternae and associated membranes (G). (C) Overview of GFP-Rab14Q70L label showing its predominant presence on EE-associated tubulo-vesicular structures, and relatively minor labeling of the Golgi region. (D) Double labeling of GFP-Rab14wt and TfR shows colocalization in narrow tubules reminiscent of recycling tubules in the vicinity of EEs (arrows) and dispersed throughout the cytoplasm (arrowheads). (E) GFP-Rab14wt partially colocalizes with syntaxin13 (stx13) in 50- to 60-nm tubules (arrows) near endosomes. (F) Similarly to Rab14wt, GFP-Rab14Q70L colocalizes with TfR in small tubules and vesicles (arrows) near EEs and scattered in the cytoplasm. No GFP label is present on lysosomes (L) or late endosomes (LE). Bars, 200 nm.
In cells transfected with the GFP-Rab14Q70L construct, the majority of the membrane-bound GFP label was again localized to narrow tubulo-vesicular structures throughout the cells and in the vicinity of EEs. The accumulation and dense clustering of these tubules around EE vacuoles was even more prominent, especially in highly expressing cells (Figure 7, C and F). Notably, a significant portion of the label in these cells was now present on the limiting membrane of the EE vacuole, with little label detectable in the Golgi region (Figure 7C).
The numerous tubulo-vesicular structures in GFP-Rab14wt– and Q70L-transfected cells morphologically resemble tubular recycling endosomes. However, in double-labeled sections of these cells, only a limited fraction of these tubules positive for Rab14wt or Q70L stained also for TfR (Figure 7, D and F) or syntaxin13 (Figure 7E). Thus, the tubulo-vesicular structures in which Rab14wt and Q70L colocalize may not (only) reflect REs, but could alternatively be transport intermediates from the TGN. However, it should be taken into account that Rab14 is highly up-regulated in overexpressing cells, such that REs “empty” with respect to cargo may be formed.
Although the bulk of the LAMP-1 label is on LE and lysosomes, where Rab14wt and Q70L are clearly absent, a minor quantity of LAMP-1 is present on tubulo-vesicular transport intermediates. In a limited fraction of these vesicles and tubules, Rab14Q70L also was present, which probably corresponds with the overlap seen in fluorescence microscopy.
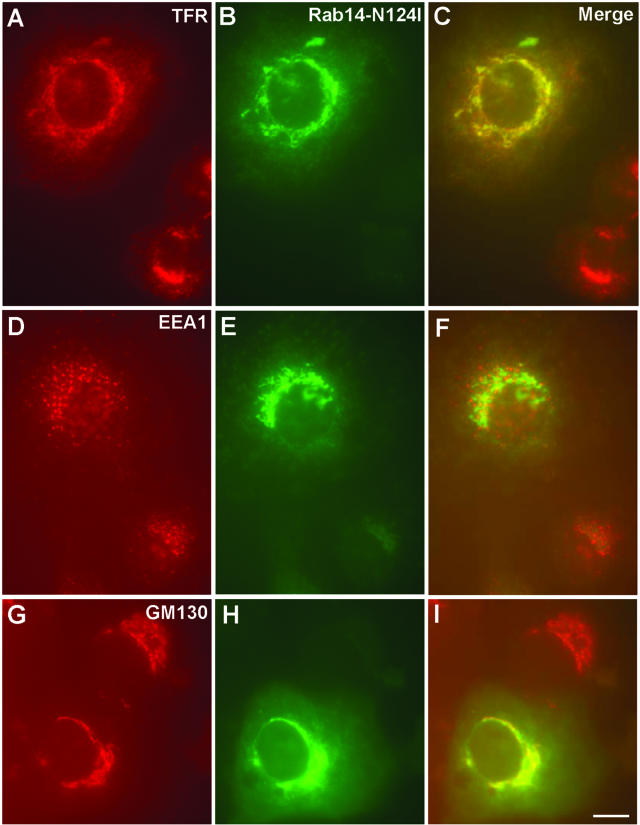
Rab14S25N and N124I Mutants Are Enriched in the Golgi Region
Immunostaining of transfected NRK cells with GM130 revealed that mutant proteins overlap well with this Golgi marker (Figure 8; our unpublished data), whereas this overlap was only partial in nontransfected or GFP-Rab14wt-transfected cells (compare Figures 3F and 5I with Figure 8I). Moreover, the distribution of TfR was changed and seemed to coincide more with Golgi stacks in the transfected cells in comparison with the nontransfected cells (Figures 3A and 8A). By contrast, there was no obvious change observed in EEA1 or LAMP-1 staining in transfected compared with nontransfected cells (Figure 8; our unpublished data).
Figure 8.
Expression of GFP-Rab14N124I mutant shifts the TfR-containing vesicles/tubules toward the Golgi region. GFP-Rab14N124I–transfected cells were stained for TfR (A), EEA1 (D), and GM130 (G) (compartments) as shown in Figure 5. Green corresponds to the GFP signal (B, E, and H). (C, F, and I) Merged images, where yellow indicates colocalization. Bars, 10 μm.
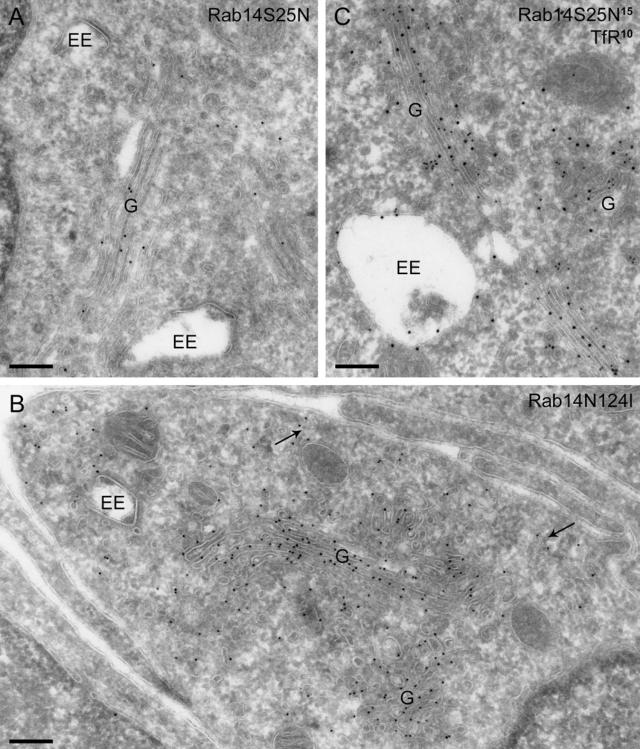
In agreement with the fluorescence microscopy data, immunoelectron microscopy showed that the major portion of Rab14S25N was in the Golgi area, mainly on the stack itself and to a lesser extent on vesicles and tubules in the vicinity (Figure 9A). Also the rough endoplasmic reticulum (RER) and the nuclear envelope contained high levels of label. Substantial label was still present on dispersed narrow tubulo-vesicular structures. In clear contrast with Q70L-expressing cells, the EE vacuoles now bore little or no label, and the clustered tubules and vesicles associated with the endosomal vacuoles were absent. The contrasting distribution patterns of the Rab14S25N versus the Q70L construct are numerically expressed in Table 1. In cells transfected with the GFP-Rab14N124I construct, the GFP label was distributed similarly to the Rab14S25N mutant, i.e., with the majority of the label in the Golgi region and little on endosomal compartments (Figure 9B).
Figure 9.
Inactive Rab14 shifts the labeling pattern toward the Golgi complex. (A) GFP-Rab14S25N, labeled with anti-GFP (10-nm gold), is mainly present on the Golgi complex (G) and largely absent from tubulo-vesicular structures near EEs. (B) Similar distribution is observed in cells transfected with GFP-Rab14N124I (10-nm gold). Some label is also present on small, dispersed vesicles (arrows). (C) Double labeling of GFP-Rab14S25N and TfR shows that the only significant overlap is in the Golgi complex, with virtually no colocalization in tubules or vesicles near endosomes. Bars, 200 nm.
Overexpression of Rab14S25N and N124I Mutants Shifts the Distribution of TfR
Double labeling with TfR showed that the major overlap between Rab14S25N, or Rab14N124I, and TfR-positive structures was in biosynthetic compartments, i.e., mainly in the Golgi area (Figure 9C). To assess whether the TfR distribution was influenced by the overexpression of the different Rab14 mutants, the distribution of the gold particles labeling the TfR was determined on randomly selected profiles of cells transfected either with Rab14S25N or with Rab14Q70L. This analysis showed that the percentage of total TfR label in Golgi-associated vesicles is moderately but significantly higher in Rab14S25N than in Q70L-transfected cells (12 vs. 5%, p < 0.01). In contrast, the fraction of TfR label distributed over small (40- to 60-nm-diameter), dispersed, and EE-associated vesicles and tubules was lower in the Rab14S25N than in the Rab14Q70L cells, i.e., 39 vs. 52% (p < 0.01). These results indicate that there is a modest net shift of TfR from the Golgi region in Rab14S25N-transfected cells toward small tubulo-vesicular structures present in a more dispersed location in the Rab14Q70L-transfected cells. The latter structures have a morphological appearance similar to vesicular transport intermediates and recycling tubules, but they are clearly distinct from EE vacuoles.
DISCUSSION
In this article, we have analyzed the tissue-specific expression pattern and intracellular localization of Rab14 to gain insight into its function. Our reverse transcription-PCR analysis for the Rab14 transcript indicated that Rab14 is expressed in all tissue types, in agreement with previous Northern blot data (Elferink et al., 1992). Western blotting analysis with newly generated, affinity-purified Rab14 antibodies paralleled the reverse transcription-PCR analysis pattern and detected a single 24-kDa band in various tissue samples. In heart tissue only, a 28-kDa band was observed, which is probably due to posttranslational modifications, because the size of the Rab14 PCR product from this tissue was unchanged. The broad tissue distribution suggests that Rab14 probably mediates a fundamental membrane-trafficking event common to all cell types. Some tissues (intestine, muscle, and thymus) express relatively low levels of Rab14, so perhaps functionally redundant isoforms substitute for Rab14 in these tissues. Alternatively, it is possible that tissues with high Rab14 levels (brain, heart, kidney, lung, placenta, and testis) might have a more prominent trafficking step mediated by Rab14.
Amino acid sequence alignment of Rab proteins surprisingly revealed that the closest homologues of Rab14 are Rab2 and Rab4, with 57 and 58% identity, respectively. Notably, Rab2 is localized to pre-Golgi intermediates termed VTCs and functions in the ER-Golgi retrograde transport pathway (Tisdale et al., 1992), whereas Rab4 is localized to endosomes and regulates the transport from EEs to REs (van der Sluijs et al., 1992). In agreement with both of these homologues, our immunofluorescence and immunoelectron microscopic studies revealed that endogenous Rab14 is localized to biosynthetic (ER, Golgi, and TGN) as well as endosomal compartments. This subcellular localization indicates that Rab14 probably regulates the transport of carrier membranes between these two sets of compartments. Previous studies on the function of several Rab proteins made use of constructs fused to the C terminus of GFP without affecting intracellular localizations (Bucci et al., 2000; Sönnichsen et al., 2000; Wang and Hong, 2002). We found that GFP-Rab14wt had a similar localization pattern to the endogenous protein, although with considerably less staining at the plasma membrane. It is possible that the marked presence of endogenous Rab14 at plasma membrane is due to another Rab14 isoform, which is not detected with the anti-GFP antibody in the overexpressing cells. In this respect it is interesting to note that a recent report showing endothelin-1 differentially regulates the expression of two Rab14 isoforms (full-length Rab14 and N-terminally truncated short Rab14) in lung fibroblast cells as established by using proteomic methods (Predic et al., 2002). Indeed, we do observe a weak ∼22-kDa band by Western blotting of rat brain and HeLa cell extracts, but only upon loading more than 50 μg of total PNS.
It has been established for several well characterized Rabs (Rab1, Rab2, Rab4, Rab5, Rab6, Rab7, and Rab9) that the Gln to Leu mutation in the PM3 motif results in a constitutively active GTPase and causes a dominant active (GTP-bound) phenotype. Conversely, the Ser/Thr to Asn mutation in the PM1 motif turns the GTPase into an inactive (GDP bound) form, resulting in a dominant negative phenotype. Based on the general principle that Rab proteins interact with their effector proteins only after GDP-to-GTP exchange, it is likely that the GTP-bound Rab mutants accumulate on their target membrane domains in association with their effectors. For example, in agreement with its role in the formation of recycling tubules, Rab4 is in its GDP-locked form concentrated near EEs, and in the GTP-locked form toward the TGN and the cell periphery. Concomitantly, the TfR is partially redistributed from an endosomal to a TGN and peripheral localization (de Wit et al., 2001). This is in line with the postulated role of Rab4 in the recycling of TfR from EEs via pericentriolar recycling compartments to the cell periphery (Daro et al., 1996). Interestingly, in comparison with Rab14wt, overexpression of Rab14Q70L significantly enhanced the presence of Rab14 in EEs and nearby tubules with a concomitant decrease in the Golgi stack. A reverse effect was seen in the case of the dominant negative mutants (S25N and N124I). Thus, the shift in distribution of the Rab14GDP- and GTP-locked forms is almost exactly the opposite as that seen for Rab4. Our observations may thus indicate that Rab14 is involved in a transport pathway from TGN to EEs. Another argument in favor of a role of Rab14 in TGN-to-EE transport is the observation that a fraction of the cell's TfR is shifted from the TGN toward a peripheral localization upon transfection with Rab14Q70L. The latter effect of Rab14 on TfR distribution is however moderate and probably indirect, consistent with an effect on newly synthesized TfR and/or the small pool of internalized TfR that is known to recycle via the TGN (Stoorvogel et al., 1988).
A small fraction of Rab14wt and Q70L colocalizes with TfR and syntaxin 13, which points to a possible function of Rab14 in a recycling pathway from EEs. If this were the case, one would expect to see enhanced formation of tubules extending from EEs as was previously reported for the Rab4 mutant. However, in spite of the accumulation of Rab14wt- and Q70L-positive tubules near EEs, the number of continuities between these tubules and the EE vacuolar membrane is not obviously augmented, thus Rab14 is unlikely to function in recycling from EEs. An explanation for the accumulation of vesicles/tubules in cells strongly overexpressing Rab14wt and Q70L is that the molecular machinery involved in the docking and fusion of vesicles and EE vacuoles is out of balance, leading to an accumulation of intermediates in the transport process. At this stage our data do not suggest a specific mechanism.
GFP movies show that GFP-Rab14wt and -Q70L fluorescence label is on dynamic tubules and vesicles that fuse with the early endosomes, whereas in the case of the dominant negative mutants (S25N and N124I), the GFP fusion protein is mostly on Golgi-like structures that are static (supplementary data; movies 1A–1D). However, transferrin-positive structures in these cells are dynamic in nature, indicating that transferrin uptake is more or less normal in these cells. We have also tested the effect of Rab14 dominant active and inactive mutants on transferrin uptake and recycling kinetics by light microscopy analysis and by fluorescence-activated cell sorting sorting GFP-Rab14 cells for internalized Alexa 594/647-transferrin. We observe little or no effect (<10% compared with control GFP-transfected cells) in Rab14Q70L- and S25N-transfected cells (our unpublished data). On the other hand, using the same protocol, >30% decrease in the transferrin uptake for Rab5 dominant inactive mutant (S34N or N133I)-transfected cells was observed, as reported previously (Gorvel et al., 1991; Bucci et al., 1992; Vitelli et al., 1997; our unpublished data).
Based on our morphological data and in comparison with Rab4 phenotypes, we propose that Rab14 may regulate the TGN to the EE/RE transport pathway. Further experiments involving the isolation of Rab14-binding proteins will be required to understand the molecular mechanism of action for this GTPase in intracellular membrane trafficking.
Supplementary Material
Acknowledgments
We thank Drs. Cary D. Austin and Suzie J. Scales for critical reading of the manuscript. We thank René Scriwanek and Marc van Peski for excellent photographic work.
Article published online ahead of print. Mol. Biol. Cell 10.1091/mbc.E03–10–0777. Article and publication date are available at www.molbiolcell.org/cgi/doi/10.1091/mbc.E03–10–0777.
Abbreviations used: EE, early endosome; EEA1, early endosomal antigen 1; GST, glutathione S-transferase; PNS, postnuclear supernatant; RE, recycling endosome; RER, rough endoplasmic reticulum; TfR, transferrin receptor; VTC, vesiculo-tubular cluster.
Online version of this article contains supporting material. Online version is available at www.molbiolcell.org.
References
- Bock, J.B., Matern, H.T., Peden, A.A., and Scheller, R.H. (2001). A genomic perspective on membrane compartment organization. Nature 409, 839–841. [DOI] [PubMed] [Google Scholar]
- Bucci, C., Parton, R.G., Mather, I.H., Stunnenberg, H., Simons, K., Hoflack, B., and Zerial, M. (1992). The small GTPase rab5 functions as a regulatory factor in the early endocytic pathway. Cell 70, 715–728. [DOI] [PubMed] [Google Scholar]
- Bucci, C., Thomsen, P., Nicoziani, P., McCarthy, J., and van Deurs, B. (2000). Rab 7, a key to lysosome biogenesis. Mol. Biol. Cell 11, 467–480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cantalupo, G., Alifano, P., Roberti, V., Bruni, C.B., and Bucci, C. (2001). Rab-interacting lysosomal protein (RILP): the Rab7 effector required for transport to lysosomes. EMBO J. 20, 683–693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carroll, K.S., Hanna, J., Simon, I., Krise, J., Barbero, P., and Pfeffer, S.R. (2001). Role of Rab9 GTPase in facilitating receptor recruitment by TIP47. Science 292, 1373–1376. [DOI] [PubMed] [Google Scholar]
- Chen, Y.A., and Scheller, R.H. (2001). SNARE-mediated membrane fusion. Nat. Rev. Mol. Cell. Biol. 2, 98–106. [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., McBride, H.M., Burgoyne, R.D., and Zerial, M. (1999a). The Rab5 effector EEA1 is a core component of endosome docking. Nature 397, 621–625. [DOI] [PubMed] [Google Scholar]
- Christoforidis, S., Miaczynska, M., Ashman, K., Wilm, M., Zhao, L., Yip, S.C., Waterfield, M.D., Backer, J.M., and Zerial, M. (1999b). Phosphatidylinositol-3-OH kinases are Rab5 effectors. Nat. Cell. Biol. 1, 249–252. [DOI] [PubMed] [Google Scholar]
- Cormont, M., Mari, M., Galmiche, A., Hofman, P., and Le Marchand-Brustel, Y. (2001). A FYVE-finger-containing protein, Rabip4, is a Rab4 effector involved in early endosomal traffic. Proc. Natl. Acad. Sci. USA 98, 1637–1642. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Daro, E., van der Sluijs, P., Galli, T., and Mellman, I. (1996). Rab4 and cellubrevin define different early endosome populations on the pathway of transferrin receptor recycling. Proc. Natl. Acad. Sci. USA 93, 9559–9564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Wit, H., Lichtenstein, Y., Kelly, R.B., Geuze, H.J., Klumperman, J., and van der Sluijs, P. (2001). Rab4 regulates formation of synaptic-like microvesicles from early endosomes in PC12 cells. Mol. Biol. Cell 12, 3703–3715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elferink, L.A., Anzai, K., and Scheller, R.H. (1992). rab15, a novel low molecular weight GTP-binding protein specifically expressed in rat brain. J. Biol. Chem. 267, 5768–5775. [PubMed] [Google Scholar]
- Gorvel, J.P., Chavrier, P., Zerial, M., and Gruenberg, J. (1991). rab5 controls early endosome fusion in vitro. Cell 64, 915–925. [DOI] [PubMed] [Google Scholar]
- Gournier, H., Stenmark, H., Rybin, V., Lippe, R., and Zerial, M. (1998). Two distinct effectors of the small GTPase Rab5 cooperate in endocytic membrane fusion. EMBO J. 17, 1930–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hales, C.M., Griner, R., Hobdy-Henderson, K.C., Dorn, M.C., Hardy, D., Kumar, R., Navarre, J., Chan, E.K., Lapierre, L.A., and Goldenring, J.R. (2001). Identification and characterization of a family of Rab11-interacting proteins. J. Biol. Chem. 276, 39067–39075. [DOI] [PubMed] [Google Scholar]
- Horiuchi, H., et al. (1997). A novel Rab5 GDP/GTP exchange factor complexed to Rabaptin-5 links nucleotide exchange to effector recruitment and function. Cell 90, 1149–1159. [DOI] [PubMed] [Google Scholar]
- Lindsay, A.J., and McCaffrey, M.W. (2002). Rab11-FIP2 functions in transferrin recycling and associates with endosomal membranes via its COOH-terminal domain. J. Biol. Chem. 277, 27193–27199. [DOI] [PubMed] [Google Scholar]
- Nagelkerken, B., Van Anken, E., Van Raak, M., Gerez, L., Mohrmann, K., Van Uden, N., Holthuizen, J., Pelkmans, L., and Van Der Sluijs, P. (2000). Rabaptin4, a novel effector of the small GTPase rab4a, is recruited to perinuclear recycling vesicles. Biochem. J. 346, 593–601. [PMC free article] [PubMed] [Google Scholar]
- Nielsen, E., Christoforidis, S., Uttenweiler-Joseph, S., Miaczynska, M., Dewitte, F., Wilm, M., Hoflack, B., and Zerial, M. (2000). Rabenosyn-5, a novel Rab5 effector, is complexed with hVPS45 and recruited to endosomes through a FYVE finger domain. J. Cell Biol. 151, 601–612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfeffer, S. (2003). Membrane domains in the secretory and endocytic pathways. Cell 112, 507–517. [DOI] [PubMed] [Google Scholar]
- Predic, J., Soskic, V., Bradley, D., and Godovac-Zimmermann, J. (2002). Monitoring of gene expression by functional proteomics: response of human lung fibroblast cells to stimulation by endothelin-1. Biochemistry 41, 1070–1078. [DOI] [PubMed] [Google Scholar]
- Prekeris, R., Davies, J.M., and Scheller, R.H. (2001). Identification of a novel Rab11/25 binding domain present in Eferin and Rip proteins. J. Biol. Chem. 276, 38966–38970. [DOI] [PubMed] [Google Scholar]
- Prekeris, R., Klumperman, J., Chen, Y.A., and Scheller, R.H. (1998). Syntaxin 13 mediates cycling of plasma membrane proteins via tubulovesicular recycling endosomes. J. Cell Biol. 143, 957–971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prekeris, R., Klumperman, J., and Scheller, R.H. (2000). A Rab11/Rip11 protein complex regulates apical membrane trafficking via recycling endosomes. Mol. Cell 6, 1437–1448. [DOI] [PubMed] [Google Scholar]
- Ren, M., Xu, G., Zeng, J., De Lemos-Chiarandini, C., Adesnik, M., and Sabatini, D.D. (1998). Hydrolysis of GTP on rab11 is required for the direct delivery of transferrin from the pericentriolar recycling compartment to the cell surface but not from sorting endosomes. Proc. Natl. Acad. Sci. USA 95, 6187–6192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slot, J.W., Geuze, H.J., Gigengack, S., Lienhard, G.E., and James, D.E. (1991). Immuno-localization of the insulin regulatable glucose transporter in brown adipose tissue of the rat. J. Cell Biol. 113, 123–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sönnichsen, B., De Renzis, S., Nielsen, E., Rietdorf, J., and Zerial, M. (2000). Distinct membrane domains on endosomes in the recycling pathway visualized by multicolor imaging of Rab4, Rab5, and Rab11. J. Cell Biol. 149, 901–914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoorvogel, W., Geuze, H.J., Griffith, J.M., and Strous, G.J. (1988). The pathways of endocytosed transferrin and secretory protein are connected in the trans-Golgi reticulum. J. Cell Biol. 106, 1821–1829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tisdale, E.J., Bourne, J.R., Khosravi-Far, R., Der, C.J., and Balch, W.E. (1992). GTP-binding mutants of rab1 and rab2 are potent inhibitors of vesicular transport from the endoplasmic reticulum to the Golgi complex. J. Cell Biol. 119, 749–761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ullrich, O., Reinsch, S., Urbé, S., Zerial, M., and Parton, R.G. (1996). Rab11 regulates recycling through the pericentriolar recycling endosome. J. Cell Biol. 135, 913–924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Urbé, S., Huber, L.A., Zerial, M., Tooze, S.A., and Parton, R.G. (1993). Rab11, a small GTPase associated with both constitutive and regulated secretory pathways in PC12 cells. FEBS Lett. 334, 175–182. [DOI] [PubMed] [Google Scholar]
- van der Sluijs, P., Hull, M., Webster, P., Male, P., Goud, B., and Mellman, I. (1992). The small GTP-binding protein rab4 controls an early sorting event on the endocytic pathway. Cell 70, 729–740. [DOI] [PubMed] [Google Scholar]
- Vitale, G., Rybin, V., Christoforidis, S., Thornqvist, P., McCaffrey, M., Stenmark, H., and Zerial, M. (1998). Distinct Rab-binding domains mediate the interaction of Rabaptin-5 with GTP-bound Rab4 and Rab5. EMBO J. 17, 1941–1951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitelli, R., Santillo, M., Lattero, D., Chiariello, M., Bifulco, M., Bruni, C.B., and Bucci, C. (1997). Role of the small GTPase Rab7 in the late endocytic pathway. J. Biol. Chem. 272, 4391–4397. [DOI] [PubMed] [Google Scholar]
- Wang, T., and Hong, W. (2002). Interorganellar regulation of lysosome positioning by the Golgi apparatus through Rab34 interaction with Rab-interacting lysosomal protein. Mol. Biol. Cell 13, 4317–4332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilcke, M., Johannes, L., Galli, T., Mayau, V., Goud, B., and Salamero, J. (2000). Rab11 regulates the compartmentalization of early endosomes required for efficient transport from early endosomes to the trans-Golgi network. J. Cell Biol. 151, 1207–1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang, J., Kim, O., Wu, J., and Qiu, Y. (2002). Interaction between tyrosine kinase Etk and a RUN domain- and FYVE domain-containing protein RUFY1. A possible role of ETK in regulation of vesicle trafficking. J. Biol. Chem. 277, 30219–30226. [DOI] [PubMed] [Google Scholar]
- Zerial, M., and McBride, H. (2001). Rab proteins as membrane organizers. Nat. Rev. Mol. Cell. Biol. 2, 107–117. [DOI] [PubMed] [Google Scholar]
- Zuk, P.A., and Elferink, L.A. (2000). Rab15 differentially regulates early endocytic trafficking. J. Biol. Chem. 275, 26754–26764. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.