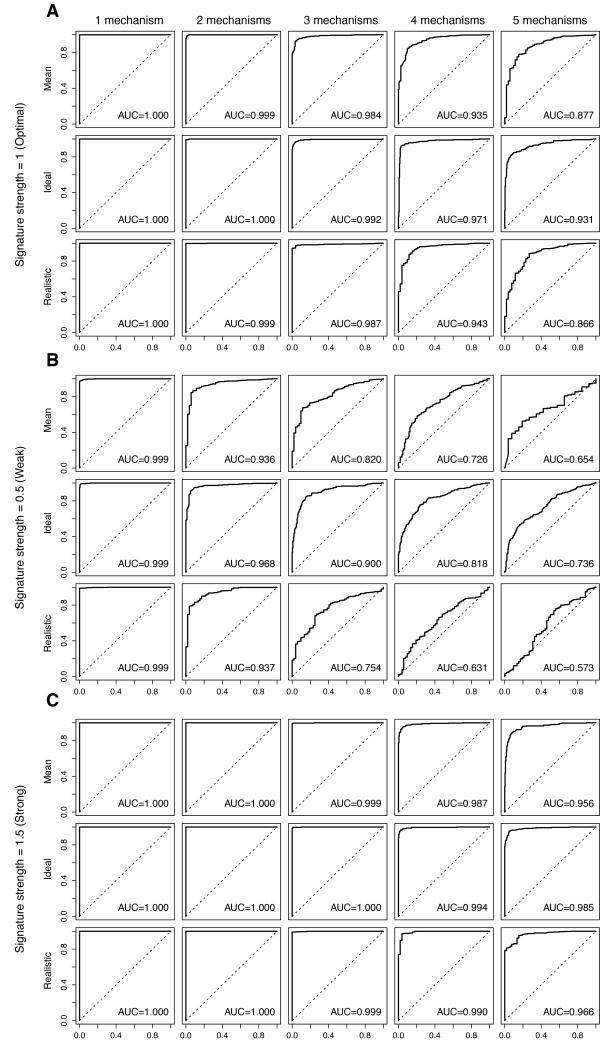
Figure 2. Impact of multiple mechanisms of resistance on the performance of the predictive signatures.
Perturbed datasets in which s% (s%=5%, 10%, 20%, 30%, 40% or 50%) of the cases were designated to be therapy sensitive were generated. Within the 1-s% resistant cases, we allocated the cases randomly into n (n=1, 2, 3, 4, 5) equally sized groups of resistance mechanisms. For each nth resistance mechanism, 100 genes were randomly selected as the “true” gene expression changes and were spiked-in by v (v=0.5, 1, 1.5). For each combination of s, n and v, we repeated the spiking and classification 100 times. Representative receiver operating characteristic (ROC) curves and the mean area under curve (AUC) for the cases are shown, where the Log2-expression of the 100-gene “true” gene expression changes were spiked-in by 1 (A, labeled “Signature strength=1 (Optimal)”), 0.5 (B, labeled “Signature strength=0.5 (Weak)”) and 1.5 (C, labeled “Signature strength=1.5 (Strong)”). Within each of A, B and C, (top row, labeled “Mean”) simulations for 1-s%=50%, 60%, 70%, 80%, 90% or 95%, (middle row, labeled “Ideal”) simulations for an optimal setting where 1-s%=50% and (bottom row, labeled “Realistic”) simulations for a clinically-realistic setting where 1-s%=90% are shown. Within each row, the representative ROCs for (from left) n=1 (“1 mechanism”), n=2 (“2 mechanisms”), n=3 (“3 mechanisms”), n=4 (“4 mechanisms”), n=5 (“5 mechanisms”) groups of distinct resistance mechanisms are shown.