Figure 2.
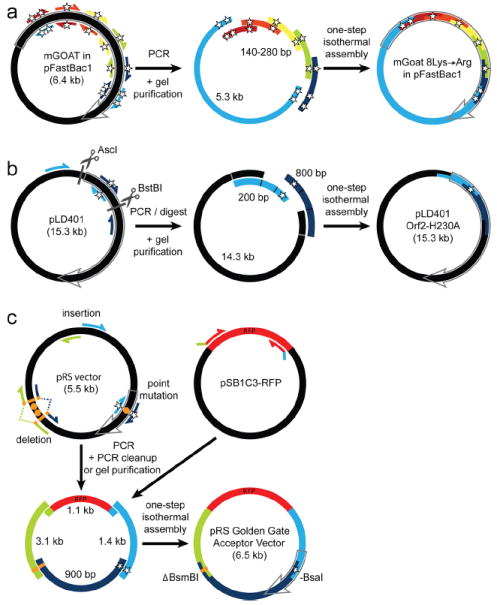
Three applications of MISO mutagenesis. (a) Simultaneous introduction of eight point mutations into the mGOAT coding sequence. Six different pairs of QuikChange-style primers were partnered (shown by colors) to generate six PCR products ranging in size from 140 bp to 5.3 kb and encoding eight lysine-to-arginine substitutions. One-step isothermal assembly reaction efficiently generated the desired lysine-free construct (see Supplementary Figure 1). (b) Introduction of a single point mutation into an existing 15.3-kb construct without vector segment amplification. QuikChange-style primers were partnered with non-mutagenic primers complementary to plasmid ends to generate two PCR products (200 bp, 800 bp). Separately, pLD401 was digested with AscI and BstBI, and the large vector fragment was gel purified. The three DNA fragments, with homologous ends, were subjected to one-step isothermal assembly to construct the mutagenized plasmid. Only the region of the plasmid produced by PCR required confirmatory resequencing. (c) Simultaneous introduction of a base substitution, deletion, and insertion into yeast shuttle vectors. One standard set of QuikChange primers (starred) plus three other primer pairs were partnered (shown by color) to generate four overlapping PCR products. One-step ISO assembly allowed deletion of two BsmBI sites from a noncoding region, recoding of one BsaI site in the bla gene, and insertion of a BsaI-flanked RFP gene to generate a new cloning site. [Open gray arrows = coding sequence; stars = point mutations; scissors = unique restriction enzyme sites; orange circles = undesirable restriction enzyme recognition sites; dashed lines = deleted region.]
