Figure 3.
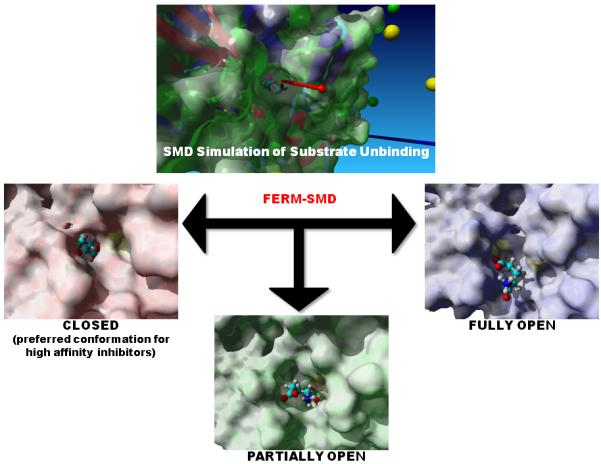
A steered molecular dynamics simulation was conducted on the glutamate-bound crystal structure of glutamate racemase from B. subtilis. A force was applied on the bound substrate along the vector indicated in the top picture (red arrow). Structures were obtained along the unbinding trajectory that correspond approximately to the following states: closed, partially open, and fully open. With substrate removed, these structures then provide the receptors for ensemble docking of the derivative library. Previous results indicate that the highest-affinity inhibitors will bind preferably to the closed conformation, over the partially and fully open conformations.
