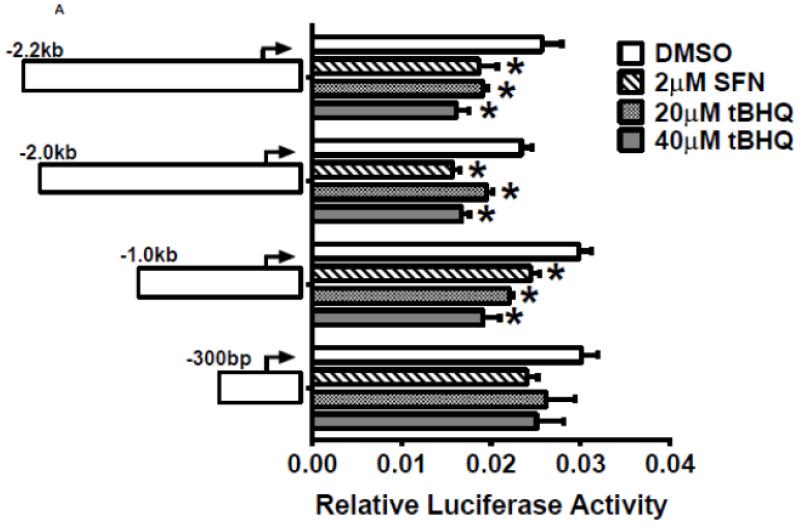
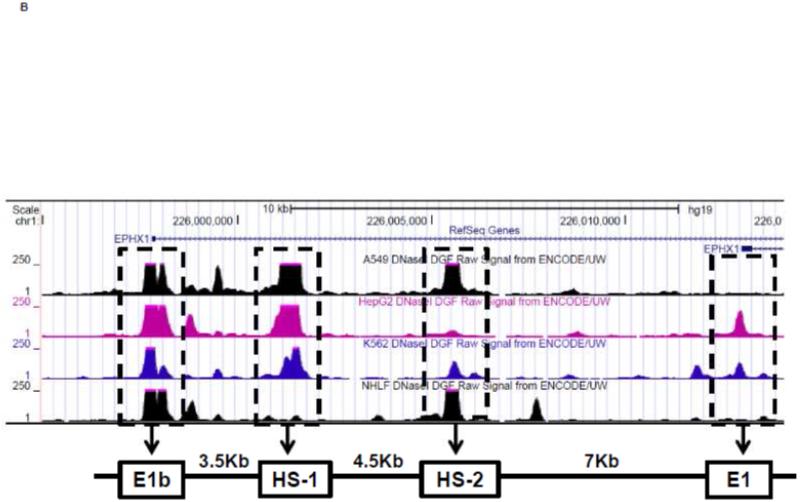
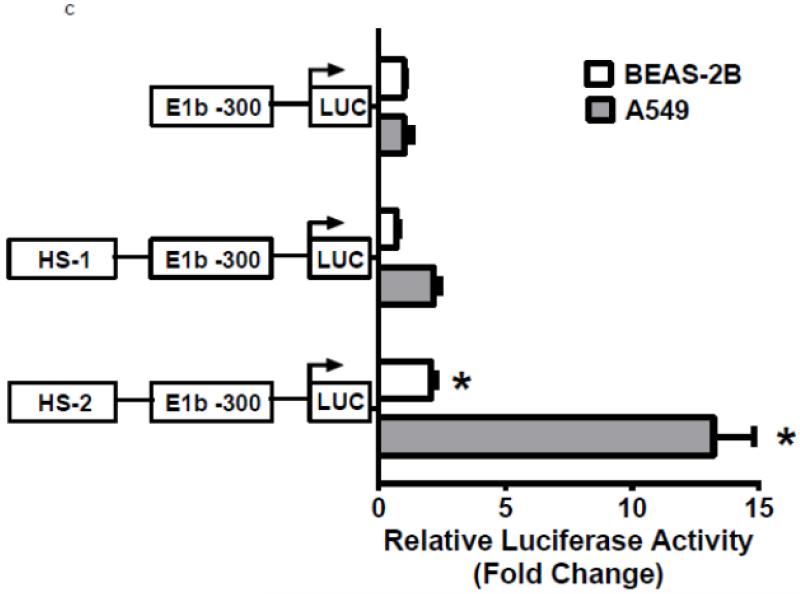
Figure 3. Identification of regulatory elements involved in E1b induction.
A.) Reporter analysis on E1b upstream sequence. BEAS-2B cells were transfected with luciferase reporter constructs driven by E1b promoters containing −300bp, −1.0kb, −2.0kb or −2.2kb promoter region. After 24h treatment with Nrf2 inducers, cells were harvested for luciferase reporter assays. Significant differences from DMSO control were indicated by “*” (p<0.05). B.) A map of the intronic region located between E1b and E1 harboring DNase hypersensitive sites in A549, HepG2, K562 and NHLF cells (modified from the UCSC genome browser). C.) Reporter analysis on DNase I hypersensitive sites. A549 and BEAS-2B cells were transfected with the E1b −300 or E1b DNase I HS/E1b-300 plasmids for 24hr and harvested for luciferase assay. Significant differences from the E1b-300 plasmid were indicated by “*” (p<0.05). All luciferase reporter activity values are expressed as the mean ± SD.