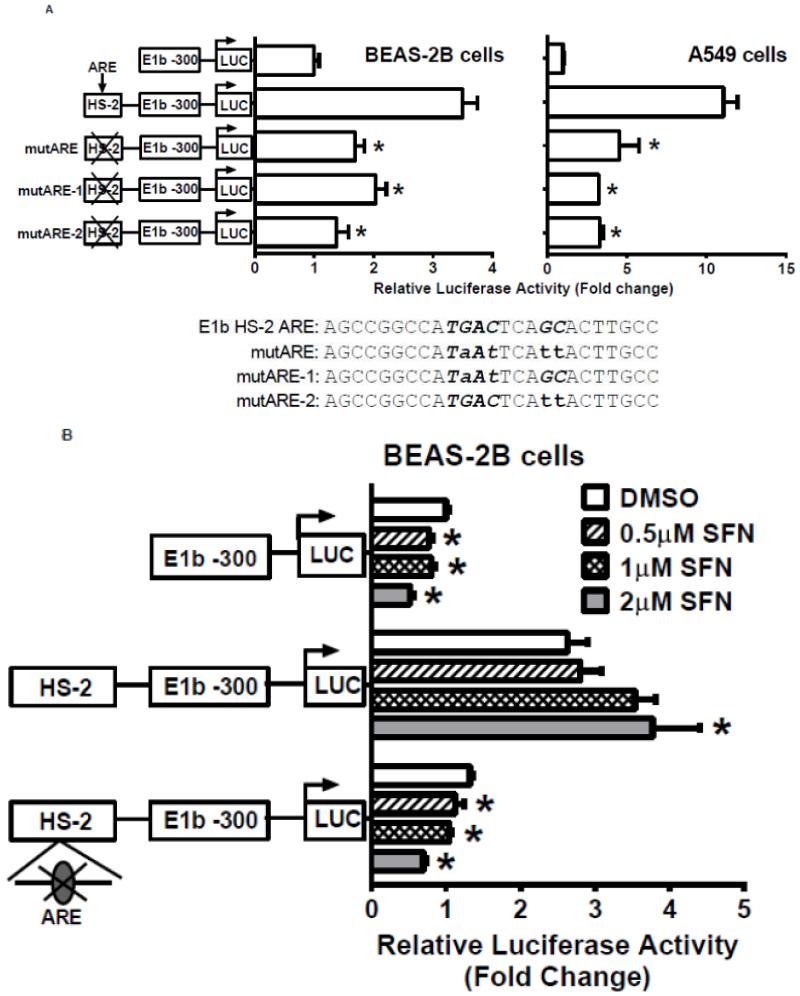
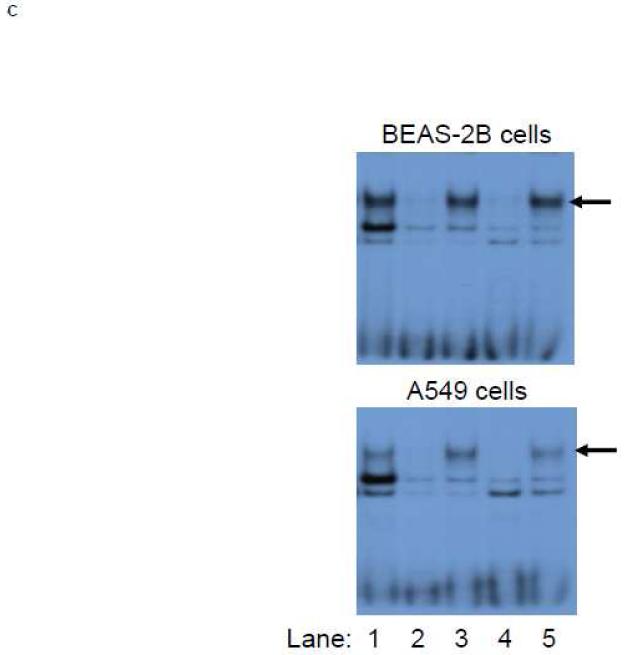
Figure 6. Identification of Antioxidant response element (ARE) within HS-2.
A.) BEAS-2B and A549 cells were transfected with E1b promoter constructs containing HS-2 (wild-type) or HS-2 with mutations in the core ARE motif. Luciferase values were determined 24 h later. *p<0.05 compared to wild-type HS-2 construct. B.) Mutation of the putative Nrf2-binding site influences SFN responsiveness of BEAS-2B cells. Cells were transfected with the reporter vectors containing E1b-300 promoter, E1b DNase I HS-2/E1b-300 promoter, or E1b DNase I mutARE/E1b-300 promoter for 18hr and treated with DMSO or SFN for 6hr before harvested for luciferase assay. All luciferase values represent mean ± SD. *p<0.05 compared to DMSO controls. C.) EMSA analysis of Nrf2 binding to the putative ARE in E1b DNase I HS-2 in SFN-treated BEAS-2B and A549 cells. E1b DNase I HS-2 ARE was end-labeled with [γ-32P]ATP and T4 kinase and incubated with nuclear extracts from untreated A549 cells or BEAS-2B cells treated with 2 M SFN for 2 h. The binding of Nrf2 to E1b DNase I HS-2 ARE was competed with 50× molar excess of cold E1b DNase I HS-2 ARE (Lane 2), mutated E1b DNase I HS-2 ARE (Lane 3), ARE from human NQO1 gene (Lane 4), or mutated NQO1 ARE (Lane 5). The arrows show specific binding.