Figure 2.
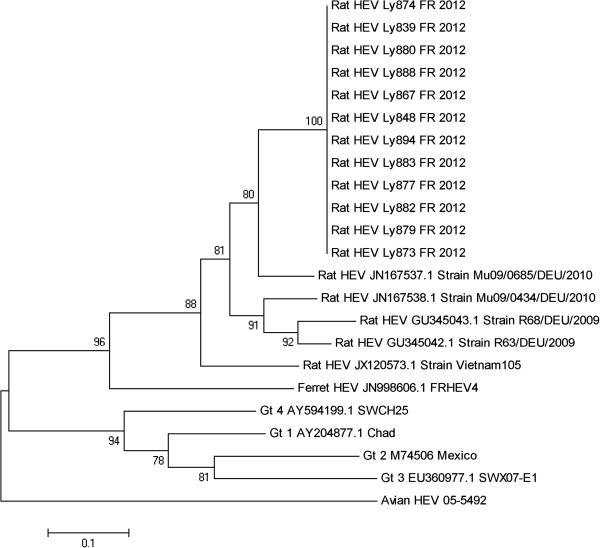
Phylogenetic tree depicting the relation between the HEV sequences from French rats to a selection of other HEV sequences. Phylogenetic tree of sequences corresponding to a 254 nt long fragment from the nested PCR product of the amplified RdRp fragment. The sequence corresponds to nucleotide position 4127 to 4371 of the rat HEV strain rat/Mu/0685/DEU2010, accession number JN167537.1. The tree was constructed by the Neighbor joining method using MEGA 5.05. The tree is depicting the relationship of the French HEV strains from 12 rats, here called “Rat HEV Ly, sample number, and Fr 2012” and described in this article, with selected HEV sequences from rat, ferret, avian HEV and genotype 1-4. The bar indicates the evolutionary distance as number of base substitutions per site. The bootstrap consensus was generated using 1000 replicates. Branches corresponding to partitions reproduced in less than 50% bootstrap replicates were collapsed. The percentage of replicate trees in which the associated taxa clustered together in the bootstrap test is shown. The tree is drawn to scale. The evolutionary distances were computed using the Tamura-Nei method (number of base substitutions per site). The rate variation among sites was modeled with a gamma distribution (shape parameter = 6). All positions containing gaps and missing data were eliminated.
