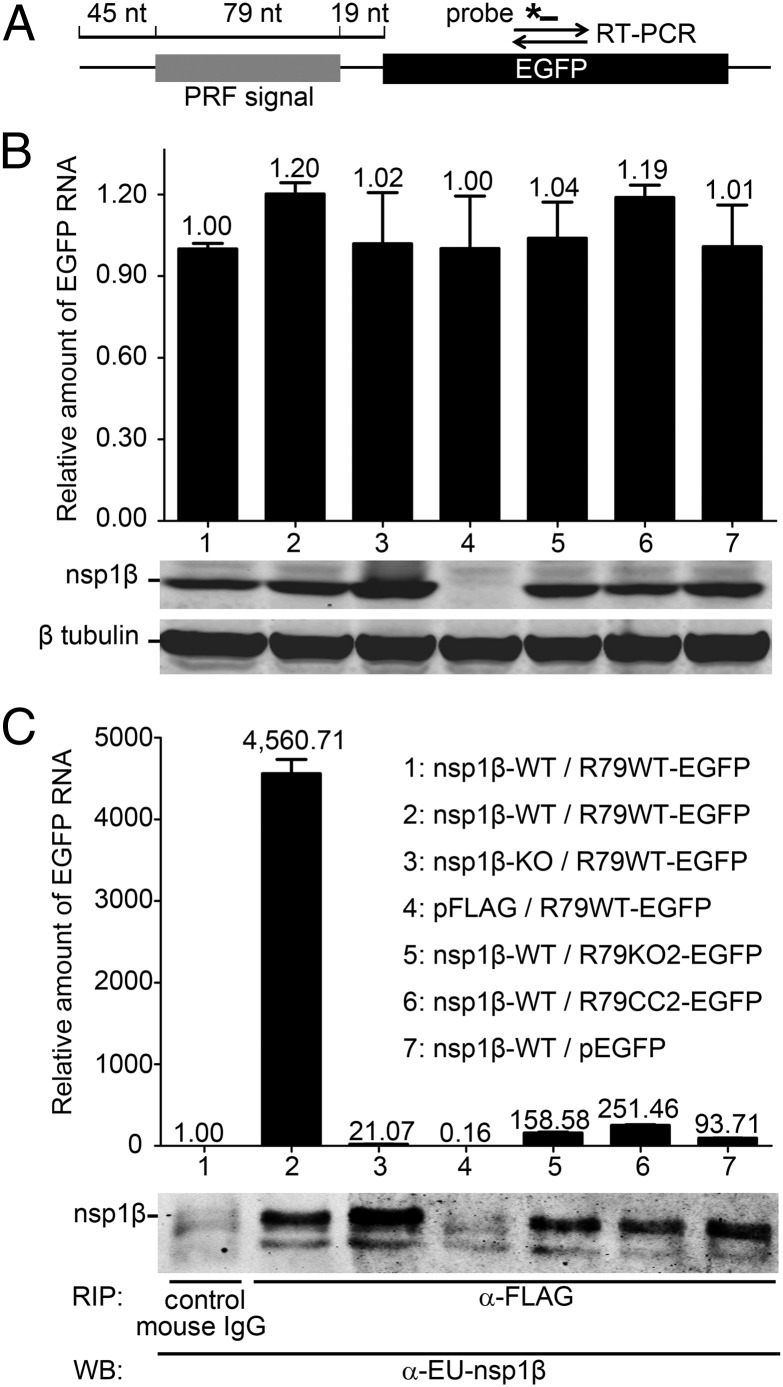
Fig. 7.
An RNA carrying the PRRSV −2/−1 PRF signal coimmunoprecipitates with nsp1β. A protein–RNA interaction assay was designed based on coimmunoprecipitation of nsp1β and RNA transcripts containing 79 nt of type 1 PRRSV sequence, including the −2/−1 PRF signal. (A) Schematic representation of the target RNA in which a WT 79-nt PRF signal (R79WT-EGFP), or its mutant KO2 or CC2 derivatives (Fig. S1A), was fused to the EGFP sequence. The latter served as a target for qRT-PCR amplification (primer set and TaqMan probe indicated), which was used to quantify the amount of RNA target bound to nsp1β. (B and C) HEK-293T cells were cotransfected with a plasmid expressing WT or 1βKO FLAG-tagged nsp1β and a plasmid expressing a WT or mutant target RNA. Empty vectors (pFLAG and pEGFP) were included as negative controls. (B) Detection of input levels of nsp1β bait and RNA target in cell lysates before the coimmunoprecipitation assay. qRT-PCR was used to determine the levels of WT or mutant R79-EGFP mRNA in transfected cells (Top). Western blot analysis was used to monitor the input of 1βKO or WT nsp1β bait (Middle) and to verify the use of equal amounts of cell lysate (β-tubulin control; Bottom). Lane numbers are explained in C. (C) Following FLAG-nsp1β immunoprecipitation, the amount of coprecipitating target RNA was determined by qRT-PCR (A). Western blot analysis using a mAb α-EU-nsp1β was used to monitor the amount of immunoprecipitated nsp1β. A legend explaining the cotransfected plasmids for each lane number is given on the right.