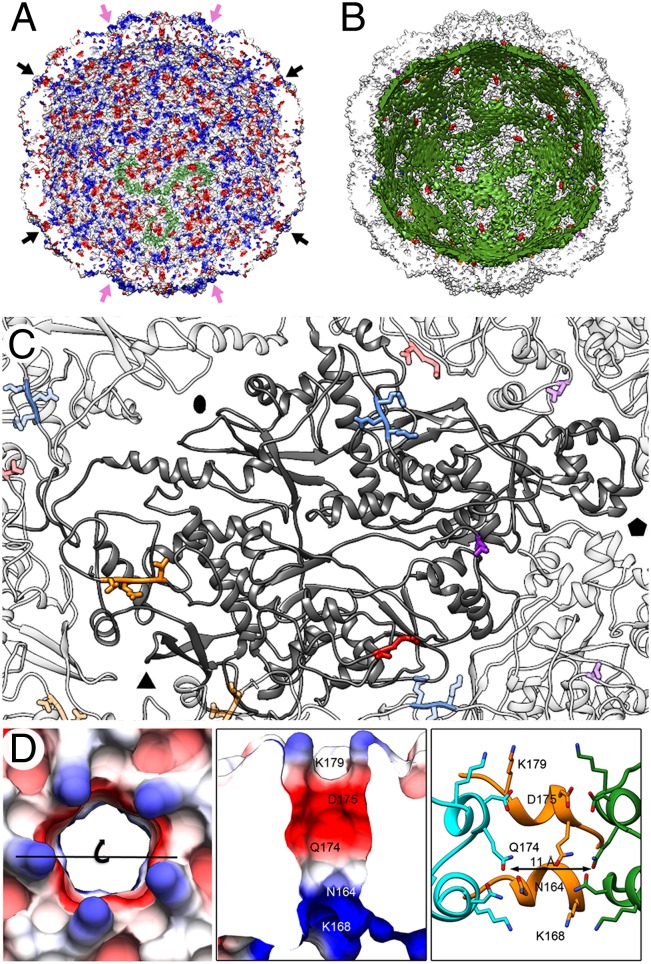
Fig. 4.
PcV CP–dsRNA interactions and pore structure. (A) PcV capsid inner surface represented with electrostatic potentials, showing the distribution of negative (red) and positive (blue) charges. The dsRNA densities have been removed computationally. Arrows indicate the capsid pores at the fivefold (black) and threefold (pink) axes. Note the triskelion-shaped electropositive areas (one is indicated in green). (B) View as for A with the PcV capsid (white) and the dsRNA outer layer (green) in close contact with the capsid inner surface (the innermost RNA shells have been stripped away). Most of three-blade–like dsRNA densities colocalize with the triskelion-shaped areas. (C) Location of the dsRNA-interacting residues in a CP subunit (dark gray): Arg67-Lys69 (blue), Leu564 (violet), Leu884-Leu887 (orange), and Leu981-Glu982 (red). Neighboring subunits and contacting residues are similarly coded in paler colors. (D) Pores at the fivefold axes showing charge distribution on the outer surface (top view, Left) and on the channel walls (slabbed side view, Center). (Right) Side view of the channel is shown, with CP subunits (green, orange and blue) represented as ribbons and the side chains exposed at the outer surface and channel wall as sticks. Note an electronegative ring toward the outer exit (built of Asp175 and Gln174) and an electropositive ring toward the inner exit (built of Asn164 and Lys168). The channel is surrounded on the outer surface by five Lys179 residues.