Fig. 3.
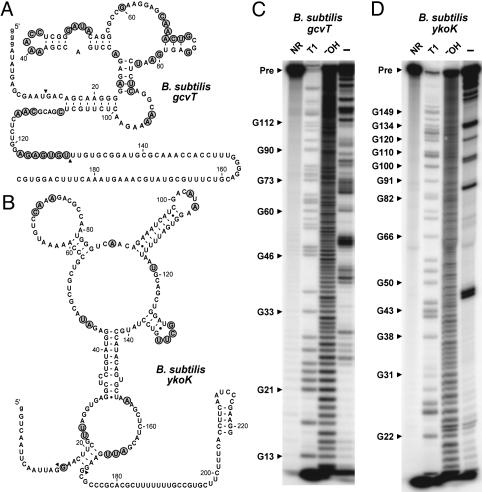
In-line probing data for the gcvT and ykoK elements from B. subtilis.(A) Sequence and secondary structure model for the 192 gcvT RNA construct. The nucleotides whose 3′ linkage undergoes a high level of spontaneous cleavage relative to most other linkages are identified by shaded circles. Arrowheads demark the boundaries of the RNA region where spontaneous cleavage data are mapped. (B) Sequence and secondary structure model for the 221 ykoK RNA construct. (C) In-line probing of 5′-32P-labeled 192 gcvT RNA. Products resulting from partial digestion with nuclease T1 (T1), partial digestion with alkali (-OH), and spontaneous cleavage during a 40-h incubation were separated by PAGE. NR, no reaction; Pre, the full-length 192 gcvT RNA. Bands corresponding to certain T1-dependent (G-specific) cleavage products are identified. Nucleotide numbering does not include the unnatural G residues (lowercase letters) inserted to improve transcription in vitro. (D) In-line probing analysis of 5′-32P-labeled 221 ykoK RNA.