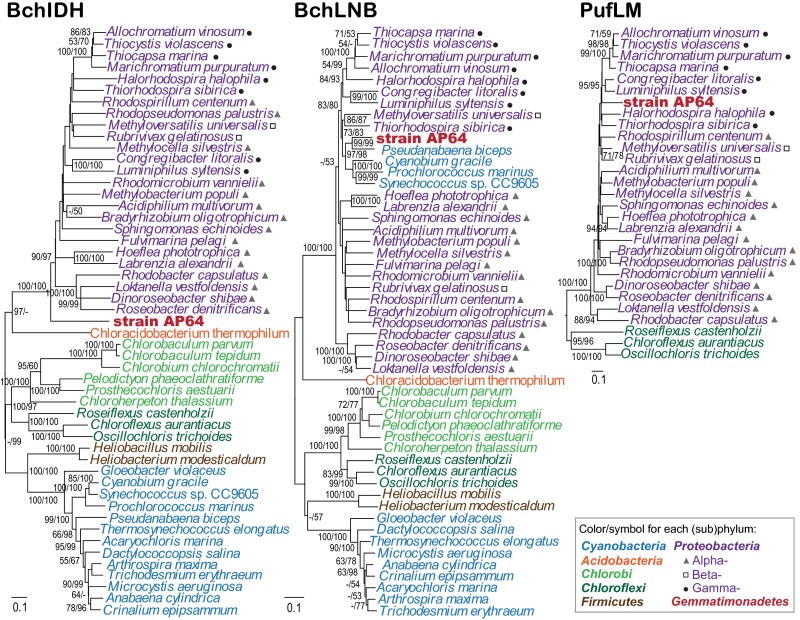
Fig. 5.
Phylogenetic analyses of AP64 photosynthesis genes. (Left ) Analysis based on concatenated alignments of amino acid sequences of magnesium chelatase (BchIDH/ChlIDH; 1,810 common amino acid positions). (Center) Analysis based on concatenated alignments of amino acid sequences of light independent protochlorophyllide reductase (BchLNB/ChlLNB; 921 common amino acid positions). (Right) Analysis based on concatenated alignments of amino acid sequences of the photosynthetic reaction center subunits (PufLM; 507 common amino acid positions). Maximum likelihood (ML) and neighbor-joining (NJ) trees were inferred for strain AP64 and representative species from other six phototrophic phyla. The PufLM tree was rooted by Chroococcidiopsis thermalis (Cyanobacteria) D1/D2 sequences. ML/NJ bootstrap values >50% are shown on the trees. Scale bars represent changes per position.