Abstract
Functional genomics studies have led to the discovery of a large amount of non-coding RNAs from the human genome; among them are long non-coding RNAs (lncRNAs). Emerging evidence indicates that lncRNAs could have a critical role in the regulation of cellular processes such as cell growth and apoptosis as well as cancer progression and metastasis. As master gene regulators, lncRNAs are capable of forming lncRNA–protein (ribonucleoprotein) complexes to regulate a large number of genes. For example, lincRNA-RoR suppresses p53 in response to DNA damage through interaction with heterogeneous nuclear ribonucleoprotein I (hnRNP I). The present study demonstrates that hnRNP I can also form a functional ribonucleoprotein complex with lncRNA urothelial carcinoma-associated 1 (UCA1) and increase the UCA1 stability. Of interest, the phosphorylated form of hnRNP I, predominantly in the cytoplasm, is responsible for the interaction with UCA1. Moreover, although hnRNP I enhances the translation of p27 (Kip1) through interaction with the 5′-untranslated region (5′-UTR) of p27 mRNAs, the interaction of UCA1 with hnRNP I suppresses the p27 protein level by competitive inhibition. In support of this finding, UCA1 has an oncogenic role in breast cancer both in vitro and in vivo. Finally, we show a negative correlation between p27 and UCA in the breast tumor cancer tissue microarray. Together, our results suggest an important role of UCA1 in breast cancer.
Keywords: p27 (Kip1), post-transcriptional regulation, lncRNA, UCA1, hnRNP I
It is well known that protein-coding genes account for only about 2% of the human genome, whereas the majority of transcripts consist of the non-coding RNAs, including microRNAs and long non-coding RNAs (lncRNAs).1 LncRNAs are transcribed mainly by RNA polymerase II; they are spliced products via canonical genomic splice site motifs, frequently ended with a poly A tail. Moreover, they are often regulated by well-established transcription factors and are expressed in a tissue-specific manner.2 Emerging evidence suggests that lncRNAs may serve as master gene regulators capable of controlling protein-coding and non-coding genes and as such they have been implicated in the regulation of a variety of cellular functions and disease processes including stemness and cancer metastasis.3, 4, 5, 6, 7, 8, 9 Various mechanisms have been proposed to explain lncRNA-mediated gene expression, and one of the important ones is their ability to form ribonucleoprotein complex through interaction with various proteins.10, 11
In support of this notion, a number of proteins have been shown to interact with lncRNAs. For example, HOTAIR is one of the first identified lncRNAs and has a critical role in cancer through epigenetic regulation mechanisms by interaction with the polycomb repressive complex 2 (PRC2),12 through which it can regulate a large number of genes. PCAT-1 is another lncRNA that has been shown to interact with PRC2 and functions as a transcriptional repressor implicated in a subset of prostate cancer patients.13 LncRNAs also interact with transcription factors to regulate gene expression. For example, PANDA interacts with the transcription factor NF-YA to limit the expression of pro-apoptotic genes; PANDA depletion markedly sensitizes human fibroblasts to apoptosis by doxorubicin.5 We have recently shown that heterogeneous nuclear ribonucleoprotein I (hnRNP I) is able to interact with lincRNA-RoR, leading to the suppression of p53 after DNA damage.14 A major role of hnRNP I is involved in mRNA splicing.15 However, hnRNP I can also serve as a translational activator by binding to 5′-untranslated region (UTR) in such a way that it recruits ribosome protein to initiate translation of several genes. For example, previous studies suggest that the 5′-UTR of p53 mRNA may form a secondary structure to serve as an internal ribosome entry site (IRES);16, 17 hnRNP I is capable of binding to the IRES to stimulate p53 translation after DNA damage, leading to the production of the N-terminal truncated isoform of p53.18 Furthermore, hnRNP I enhances the IRES-dependent translation of p27 (Kip1) mRNA and modulates transition from the G1 to S phase.19
p27 is a well-known tumor suppresser because it has a role in the suppression of cyclin-dependent kinase.20 p27 is frequently downregulated in cancer including breast cancer;21 the reduced expression of p27 protein is associated with poor clinical outcome of breast cancer, and thus is implicated in cell proliferation and differentiation.22 The regulation of p27 is complex, involving transcriptional, translational and post-translational levels.23, 24 However, it is not clear whether the p27 protein level can be regulated by lncRNAs.
Urothelial carcinoma-associated 1 (UCA1) is a lncRNA originally identified in bladder transitional cell carcinoma.25 The entire sequence consists of three exons with 1.4 kb in length. As it is highly expressed in bladder transitional cell carcinoma, it was suggested to serve as a biomarker for the diagnosis of bladder cancer. Subsequently, another isoform (2.2 kb) was identified by a different group as cancer upregulated drug resistant (CUDR) gene in the doxorubicin-resistant subline of human squamous carcinoma A431 cells.26 Sequence alignment revealed that the 1.4 isoform is contained in the 2.2 kb isoform. Although these studies suggest an oncogenic role of UCA1 in bladder cancer, the underlying mechanism is largely unknown. Moreover, the role of UCA1 in other type of cancers such as breast cancer remains to be determined.
In the present study, we show that UCA1 promotes breast cancer cell growth in both in vitro and in vivo. Importantly, our results suggest that UCA1 is able to suppress the tumor suppressor p27 through interaction with hnRNP I. We also show that there is a negative correlation between UCA1 and p27 in breast cancer specimens, highlighting its clinical significance.
Results
Identification of UCA1 as an hnRNP I-binding partner
We have previously identified hnRNP I as a binding partner of lincRNA-RoR (RoR) by RNA precipitation combined with proteomics analysis.14 Although hnRNP I promotes p53 translation through interaction with the 5′-UTR of p53 mRNA, RoR may interrupt this interaction so that RoR represses p53 in response to DNA damage.14 As an RNA-binding protein, hnRNP I has multiple functions and has been implicated in diverse cellular processes such as alternative splicing, neural development, neural transdifferentiation and tumorigenesis.27, 28, 29, 30 Thus, we asked whether hnRNP I can functionally interact with other lncRNAs in addition to RoR. Toward this end, we performed RNA immunoprecipitation (RIP) assays with hnRNP I antibody and then profiled lncRNA expression using the RT–PCR-based Human Disease-related LncRNA Profiler which carried primer sets of 83 lncRNAs mainly derived from lncRNA database.31
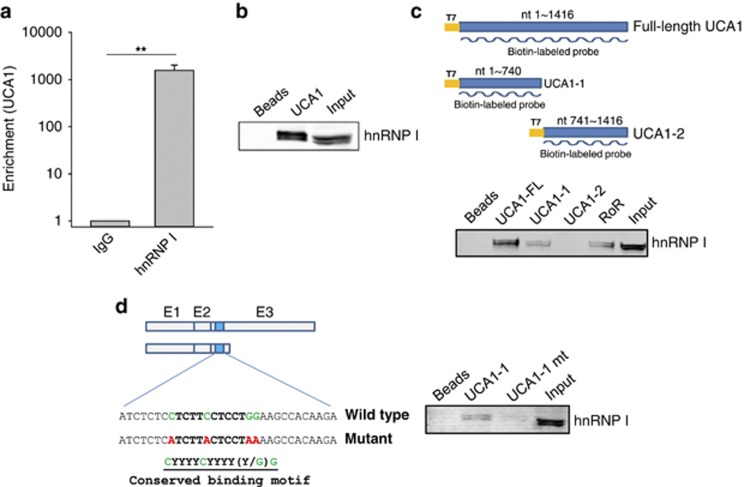
As expected, RoR was able to interact with hnRNP I. Importantly, we identified additional 18 lncRNAs (Supplementary Figure S1). Among them, UCA1 was on the top of the list in terms of enrichment. Thus, we selected UCA1 for further characterization. To verify its interaction with UCA1, we designed a different set of UCA1 primers (UCA1-RT-5.1 and UCA1-RT-3.1) and detected over a 1000-fold enrichment of UCA1 by hnRNP I antibody compared with IgG control (Figure 1a). To further confirm this interaction between hnRNP I and UCA1, we performed RNA precipitation using the synthetic biotin-labeled UCA1 RNA probe. Three UCA1 isoforms have been reported; they are 1.4, 2.2 and 2.7 kb in length.25, 26 Although the 1.4 and 2.2 kb isoforms have been characterized (Supplementary Figure S2), little is known about the 2.7 kb form. As the 1.4 kb form is included in the 2.2 kb form except the first 14 nucleotides, we made a biotin-labeled RNA probe based on the 1.4 kb isoform, using T7 RNA polymerase. After RNA precipitation, western blot revealed an hnRNP I-specific doublet band (Figure 1b).
Figure 1.
Identification of UCA1 as an hnRNP I-binding partner. (a) Enrichment of UCA1 by hnRNP I antibody, as detected by RIP assay. (b) Confirmation of the interaction between UCA1 and hnRNP I by RNA precipitation assay using UCA1 RNA probe. Preparation of UCA1 RNA probe and cellular extract from MCF-7 cells were detailed in the text. (c) The 5′ end of UCA1 is responsible for its interaction with hnRNP I, as detected by RNA precipitation. UCA1 RNA probes used in this experiment were indicated in top panel. RoR exon 4 was used as a positive control. (d) A putative hnRNP I-binding motif in UCA1 is critical to its interaction with hnRNP I. Description of the mutant UCA1 at the putative hnRNP I-binding site was indicated in left panel where the conserved binding motif was underlined. Y, either C or T. Detection of hnRNP I in the pellets by western blot as indicated in right panel. Error bars represent S.E.M., n=3. **P<0.01
To determine which part of UCA1 involves the interaction with hnRNP I, we generated two separate probes covering the 5′ end and the 3′ end, respectively (Figure 1c, top panel). Although both UCA1-1 (5′ end) and the full-length UCA1 probes detected the hnRP I signal, UCA1-2 (3′ end) did not reveal any visible signal (Figure 1c, bottom panel), suggesting that the first part of UCA1 is responsible for the interaction with hnRNP I. hnRNP I is also named polypyrimidine tract-binding protein 1 (PTBP1), meaning that it can bind to a pyrimidine-rich region of RNA. Thus, we scanned the first part of UCA1 and found a sequence 5′-CTCTTCCTCCTGG-3′ around nucleotide 700 which was within UCA1-1. This sequence is very similar to the conserved hnRNP I-binding motif 5′-CYYYYCYYYY(Y/G)G-3′ where Y stands for C or T.32 Therefore, we mutated this site (Figure 1d, left panel) and found that the interaction was substantially reduced (Figure 1d, right panel), suggesting that this hnRNP I-binding motif is critical to their interaction.
hnRNP I stabilizes UCA1 RNA
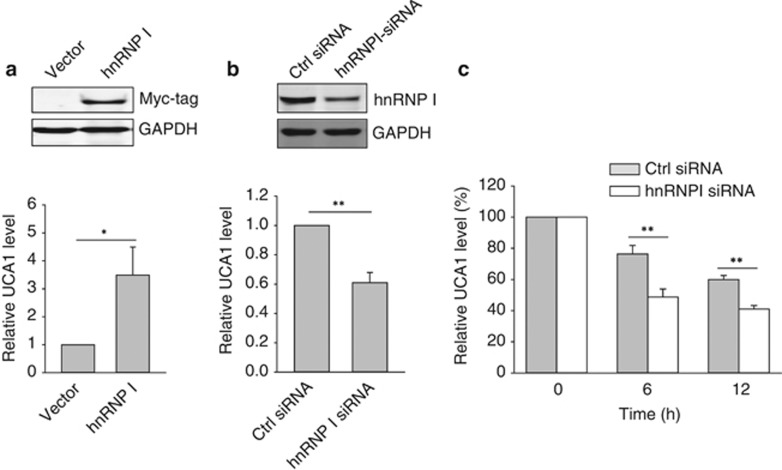
To characterize the function of this interaction between hnRNP I and UCA1, we examined the effect of hnRNP I on UCA1 expression. Ectopic expression of hnRNP I increased the UCA1 level (Figure 2a). We also knocked down hnRNP I by RNAi and found that hnRNP I-siRNA reduced the UCA1 level (Figure 2b), suggesting that UCA1 is regulated by hnRNP I. As one function of hnRNP I is able to regulate RNA stability,33 we examined whether hnRNP I can regulate UCA1 RNA stability. MCF-7 cells were first transfected with control siRNA or hnRNP I-siRNA, and then treated with 1 μg/ml of actinomycin D.33 Total RNA was isolated 6 or 12 h after the treatment. As shown in Figure 2c, the UCA1 level was significantly lower in the hnRNP I-siRNA cells than in control siRNA cells at both 6 and 12 h time points, suggesting that hnRNP I can increase the UCA1 stability.
Figure 2.
Effect of hnRNP I on UCA1 expression. (a) Ectopic expression of hnRNP I increases UCA1, as detected by qRT–PCR. (b) Suppression of UCA1 by hnRNP I-siRNA. (c) Effect of hnRNP I-siRNA on UCA1 stability. MCF-7 cells were first transfected with control siRNA or hnRNP I-siRNA overnight and the transfected cells were treated with actinomycin D at 1 μg/ml. RNA was isolated at 0, 6 and 12 h, respectively. Error bars represent S.E.M., n=3. *P<0.05; **P<0.01
Doxo induces the UCA1 level by increasing the interaction between UCA1 and hnRNP I
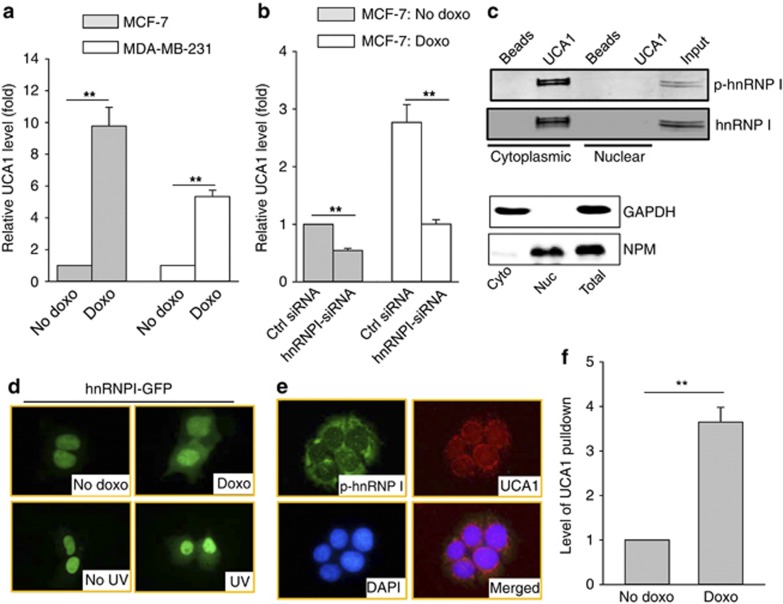
UCA1 has been implicated in drug resistance. In particular, the 2.2 kb isoform was isolated from the doxorubicin (doxo) resistant cell lines.26 Although we found that UCA1 did not significantly impact the response of MCF-7 cells to doxo (not shown), doxo was able to induce UCA1 by about a 10-fold in MCF-7 cells (Figure 3a). As doxo is a well-known p53 inducer, to determine whether doxo-induced UCA1 involves p53, we did the same experiment in MDA-MB-231 cells which carry a mutant p5334 and found that doxo was still able to induce UCA1 (Figure 3a), suggesting that it is independent of p53. To better characterize this doxo-induced UCA1 expression, we treated HCT-116 p53-WT and HCT-116 p53-null cells with doxo. Evidently, there was a similar induction of UCA1 by doxo in both cell lines (Supplementary Figure S3), further supporting the p53-independent induction of UCA1 by doxo. Finally, hnRNP I-siRNA was able to block the doxo-induced UCA1 (Figure 3b), suggesting that hnRNP I is required for this induction.
Figure 3.
Induction of UCA1 through interaction with hnRNP I. (a) Doxo induces UCA1 independent of p53, as detected by qRT–PCR in MCF-7 and MDA-MB-231 cells. Cells were treated with doxo at 1 μg/ml for 24 h before they were harvested for RNA extraction. (b) Suppression of doxo-induced UCA1 by hnRNP I-siRNA. MCF-7 cells were first transfected with control siRNA or hnRNP I-siRNA and then treated with doxo at 1 μg/ml for 24 h before they were harvested for RNA extraction. (c) The phosphorylated form of hnRNP I is localized in the cytoplasm and is responsible for interaction with UCA1. RNA precipitation was used to detect hnRNP I. The same membrane was probed simultaneously with hnRNP I antibody (mouse origin) and p-hnRNP I antibody (rabbit origin), followed by secondary antibody labeled with either IRDye 680 or IRDye 800. The bottom panel shows the cytoplasmic (cyto) and nuclear (nuc) extracts used for RNA precipitation assays in the top panels. GAPDH serves as a cytoplasmic marker and NPM as a nuclear marker. (d) Redistribution of hnRNP I in response to doxo treatment. MCF-7 cells were first transfected with GFP–hnRNP I fusion construct, followed by doxo treatment (1 μg/ml) for 8 h or UV treatment (20 J/m2) for 3 h before fixation for fluorescent microscopy. (e) Colocalization of phosphorylated hnRNP I with UCA1 by IF and fluorescence in situ hybridization. (f) Doxo increases UCA1 pulldown by hnRNP I antibody. MCF-7 cells were treated with doxo the same way as in d, and then cytoplasmic fraction was prepared for RIP assay. Error bars represent S.E.M., n=3. **P<0.01
Given the ability of hnRNP I to increase the UCA1 RNA stability (Figure 2c), we then determined whether this doxo-induced UCA1 is due to the enhancement of the interaction between hnRNP I and UCA1. Cell fractionation revealed that only the cytoplasmic hnRNP I was detected in the cytoplasm (Figure 3c). Importantly, this cytoplasmic hnRNP I was phosphorylated (Figure 3c), in consistent with our previous finding.14 Of interest, doxo caused subcellular relocalization of hnRNP I, and more hnRNP I was seen in the cytoplasm after doxo treatment (Figure 3d and Supplementary Figure S4A), as detected by hnRNP I–GFP fusion protein. Similarly, UV was also able to increase cytoplasmic hnRNP I (Figure 3d). These results suggest that DNA damage causes phosphorylation of hnRNP I, leading to the cytoplasmic accumulation, and this phosphorylated form of hnRNP I is responsible for its interaction with UCA1. In support of this finding, we showed that both phosphorylated hnRNP I and UCA1 were colocalized to the cytoplasm, especially in the perinuclear region, by immunofluorescence (IF) and fluorescence in situ hybridization (FISH) (Figure 3e and Supplementary Figure S4B). Finally, immunoprecipitation with hnRNP I antibody revealed that the level of UCA1 was higher in doxo-treated cells than in no doxo cells (Figure 3f), implying that more cytoplasmic hnRNP I after doxo treatment leads to more pulldown of UCA1.
UCA1 suppresses the p27 protein level by competing with p27 mRNA for hnRNP I
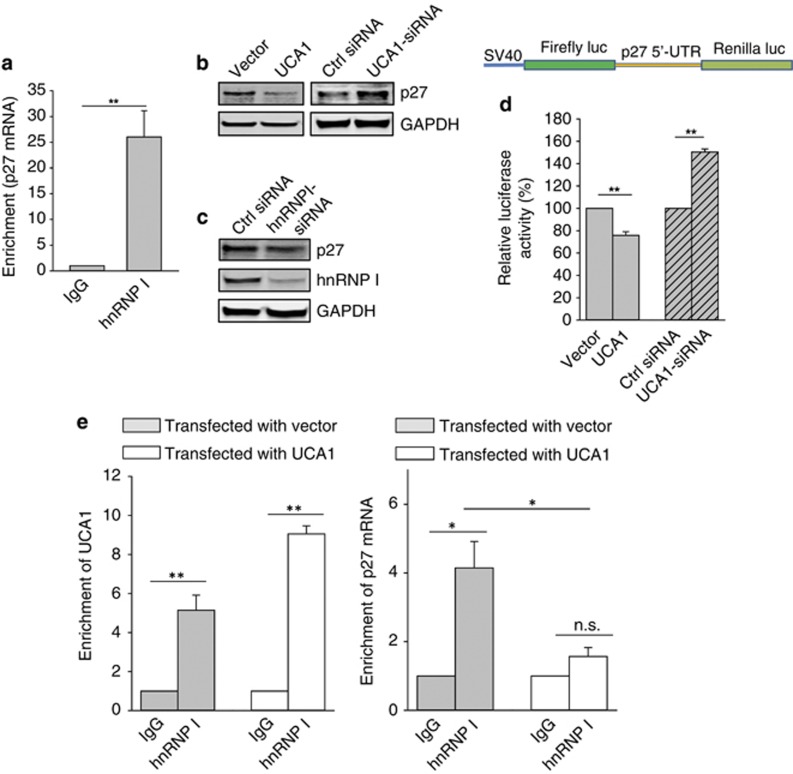
Next, we determined whether the tumor suppressor p27 serves as a potential target for UCA1 because a previous report suggested that hnRNP I interacts with p27 and enhances p27 translation through interaction with the IRES, leading to cell cycle arrest.19 RIP assay using hnRNP I antibody confirmed that hnRNP I interacted with p27 mRNA (Figure 4a). We then determined the effect of UCA1 on p27 expression. Ectopic expression of UCA1 suppressed the p27 protein level; in contrast, UCA1-siRNA increased the p27 protein level (Figure 4b). However, UCA1 had no effect on the p27 mRNA level as detected by quantitative RT–PCR (qRT–PCR) (Supplementary Figure S5), suggesting a post-transcriptional regulation mechanism involvement. To determine the role of hnRNP I in this UCA1-mediated p27 protein expression, we suppressed hnRNP I by RNAi and as expected, hnRNP I-siRNAs reduced the p27 protein level (Figure 4c). As it has been previously shown that hnRNP I interacts with the 5′-UTR of p27,19 we cloned this region into a dual luciferase reporter (Figure 4d, top panel). We were able to show that UCA1 affected the p27 5′-UTR-driven renilla luciferase activity (Figure 4d, bottom panel). For example, ectopic expression of UCA1 caused over 20% reduction of the renilla luciferase activity and UCA1-siRNA increased the activity by over 40%. Together, these results demonstrate that UCA1 is an important factor contributing to the regulation of p27 protein expression.
Figure 4.
UCA1 suppresses p27 protein level by competing for hnRNP I. (a) Interaction of p27 mRNA with hnRNP I, as detected by RIP assay. (b) Although ectopic expression of UCA1 suppresses, UCA1-siRNA increases the p27 protein level. MCF-7 cells were transfected with vector or UCA1; control siRNA or UCA1-siRNA and then harvested for western blotting 24 h after transfection. (c) Suppression of the p27 protein level by hnRNP I-siRNA. The procedure was same as in b. (d) Effect of UCA1 and UCA1-siRNA on the p27 5′-UTR luciferase reporter activity. MCF-7 cells were transfected with the p27 5′-UTR luciferase reporter along with UCA1 or UCA1-siRNA. Luciferase assay was carried out 24 after transfection. Relative luciferase activity is Renilla luciferase activity relative to Firefly luciferase activity as an internal control. (e) Suppression of the interaction of p27 mRNA with hnRNP I by UCA1. MCF-7 cells were transfected with vector control or UCA1, and cellular extract was prepared for RIP assay using hnRNP I antibody 24 h after transfection. Error bars represent S.E.M., n=3. *P<0.05; **P<0.01
To further determine how the interaction of hnRNP I with UCA1 suppresses the p27 protein level, we performed a competition assay. In these experiments, we used a smaller number of cells for RIP. As expected, we observed more UCA1 pulldown by hnRNP I antibody (approximately ninefold) for UCA1-transfected cells than for vector control cells (approximately fivefold) (Figure 4e, left panel). However, ectopic expression of UCA1 did not significantly increase the enrichment of p27 mRNA by hnRNP I antibody (IgG versus hnRNP I antibody); instead UCA1 significantly reduced the ability of hnRNP I to pull down p27 mRNA (Figure 4e, right panel), suggesting that UCA1 may compete with p27 mRNA for hnRNP I. In consistent with this finding, RNA precipitation assays with UCA1 probe revealed consistently pulldown of hnRNP I, but we were not able to detect p27 mRNA from the same pellet (Supplementary Figure S6), suggesting that, although both UCA1 and p27 mRNA can bind to hnRNP I, they are in separate complexes.
UCA1 promotes cell growth and tumorigenesis
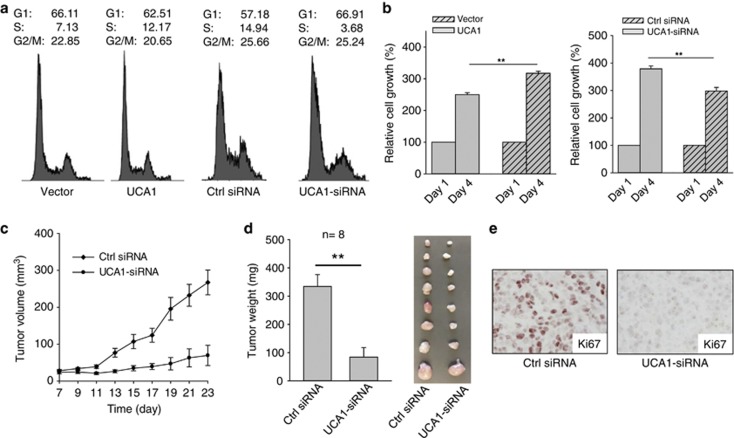
p27 is a well-known tumor suppressor that controls cell cycle transition from G1 to S.35 Thus, we determined the effect of UCA1 on cell cycle. Ectopic expression of UCA1 increased the S phase population (12.17% for UCA1 versus 7.13% for vector control); at the same time, G1 population was decreased from 66.11% in vector control to 62.51% in the UCA1 cells (Figure 5a). On the other hand, UCA1-siRNA decreased the S phase population (3.69% for UCA1-siRNA versus 14.95% for control siRNA); G1 population increased to 66.91% in the UCA1-siRNA cells from 57.18% in control siRNA (Figure 5a). The effect of UCA1-siRNA on cell cycle seems more prominent than the ectopic expression presumably because the endogenous level of UCA1 is relatively high in cancer cells.
Figure 5.
UCA1 promotes tumor cell growth. (a) Effect of UCA1 on cell cycle progression. MCF-7 cells were transfected with vector control or UCA1; control siRNA or UCA1-siRNA. The transfected cells were harvested for FACS analysis 24 h after transfection. (b) Effect of UCA1 on cell growth as detected by the MTT assay. (c and d) Effect of UCA1-siRNA on tumor growth in a xenograft mouse model. MCF-7 cells were transfected with control siRNA or UCA1-siRNA, and then injected into mammary fat pads as described in the text. Tumor growth was measured every other day after 7 days of injection (c) and tumors were then harvested at day 23 and weighed (d). Actual tumor size after harvest was shown in the right panel. (e) Detection of the cell proliferation marker Ki-67 in xenograft tumors. Error bars represent S.E.M., n=3 except for indicated. **P<0.01
Having demonstrated the functional interaction between UCA1 and hnRNP I, through which UCA1 is capable of suppressing p27, we next determined the role of UCA1 in breast cancer. As shown in Figure 5b, ectopic expression of UCA1 increased cell growth; in contrast to ectopic expression, UCA1-siRNA suppressed cell growth. For example, at day 4, relative cell growth for UCA1 cells is over 300% compared with ∼250% for the vector control. With regard to UCA1-siRNA groups, the relative cell growth at day 4 for UCA1-siRNA was ∼290% compared with ∼380% for control siRNA. To further determine the role of UCA1 on breast tumor growth, we injected MCF-7 cells transfected with either control siRNA or UCA1-siRNA into female nude mice. In consistent with in vitro results, UCA1-siRNA significantly reduced tumor growth (Figure 5c) and resulted in a reduction of tumor weight (Figure 5d). Moreover, we detected a stronger Ki-67 expression for tumors derived from control siRNA than that from UCA1-siRNA (Figure 5e and Supplementary Figure S7), further supporting the role of UCA1 in tumor cell growth and proliferation.
Upregulation of UCA1 in breast tumor specimens and its expression correlates with downregulation of p27 expression
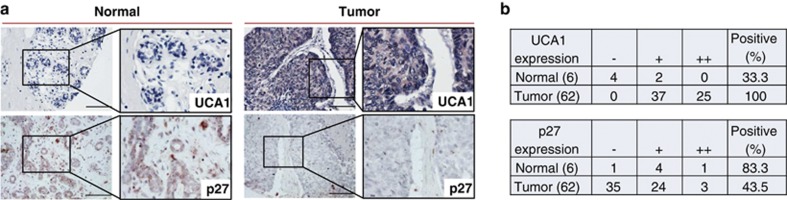
To determine the clinical relevance of this UCA1-mediated p27 expression, we detected UCA1 expression in breast cancer tissue microarrays (TMAs) by chromogenic in situ hybridization (CISH) and found that UCA1 was upregulated in breast tumors compared with the normal breast tissue (Figure 6a). In contrast, p27 was downregulated in breast tumors, which was derived from the same core (Figure 6a). This negative correlation between UCA1 and p27 was further supported by the data from breast cancer TMA. For example, among 62 tumor cores in the TMA, all of them expressed UCA1, whereas over 50% tumors revealed no p27 staining (Figure 6b). For normal tissues, there were 33.3% positive for UCA1 and 83.3% positive for p27, suggesting a clinical significance of UCA1 in breast cancer.
Figure 6.
Negative correlation between UCA1 and p27 protein in breast cancer specimens. (a) Representative pictures showing expression of UCA1 and p27 protein in the same cores. (b) Overall relative expression of UCA1 and p27 protein in the TMA
Discussion
LncRNAs have emerged as an important player in cancer biology. Increasing evidence suggests that lncRNAs may function as oncogenes or tumor suppressors. They are frequently dysregulated in cancer. In support of this notion, we show that UCA1 is upregulated in breast cancer and its expression is negatively correlated with the tumor suppressor p27. Furthermore, although ectopic expression of UCA1 promotes, UCA1-siRNA suppresses tumor cell growth in vitro and tumor growth in vivo. Importantly, our results suggest that this UCA1-mediated tumor growth is at least in part through interaction with hnRNP I, leading to the suppression of p27 protein expression. Thus, our study provides supporting evidence that UCA1 has an oncogenic role in breast cancer.
One of important findings in this study is the identification of p27 as a UCA1 target through hnRNP I. p27 is a well-known tumor suppressor in various types of cancer. In breast cancer, p27 was shown to suppress cell cycle and cause cell cycle arrest at G1.21 A low level of p27 is often associated with ER- and PR-negative breast cancer, and more likely to have a high-grade tumor.36 p27 belongs to the Cip/Kip family of cyclin-dependent kinase (Cdk) inhibitor proteins;37 it binds to cyclin D either alone or when complexed to its catalytic subunit CDK4 and prevents the activation of cyclin D–CDK4 complexes, and thus controls the cell cycle progression at G1. In consistent with this, we show that, although the ectopic expression of UCA1 decreases, UCA1-siRNA increases the G1 population. Moreover, tumors harvested from mice reveal a much weaker signal for Ki-67 in UCA1-siRNA than in control siRNA. These results suggest that UCA1 promotes tumor growth through increased cell proliferation, which may also explain in part why UCA1 is highly expressed in breast tumor specimens compared with normal breast tissues, although the analysis of a larger number of breast cancer specimens is needed to statistically demonstrate this negative correlation with p27 expression.
Our study identifies hnRNP I as an important player in this UCA1-mediated p27 repression network because the ability of UCA1 to repress p27 is dependent on the interaction of UCA1 with hnRNP I. Suppression of hnRNP I by RNAi decreases p27, as UCA1-siRNA does. In particular, we identify a potential hnRNP I-binding motifs in UCA1. This motif is critical because mutations with two Ts and two Gs substantially decrease its ability to bind to hnRNP I (Figure 1d), providing further evidence of the specific interaction between hnRNP I and UCA1. It is well known that hnRNP I is an RNA-binding protein that carries several RNA-binding domains and it has an active role in mRNA splicing.38 The level of hnRNP I is relatively high in the cell. However, the majority of hnRNP I is retained in the nucleus and this may have to do with its nuclear function as a splicing regulator. Only a small fraction of hnRNP I is in the cytoplasm and it is phosphorylated.14 Our study suggests that this cytoplasmic phosphorylated hnRNP I is responsible for the interaction with UCA1. This is not surprising because protein translation takes place in the cytoplasm. Of interest, doxo can increase the cytoplasmic localization of the phosphorylated hnRNP I. When more phosphorylated hnRNP I is accumulated in the cytoplasm, we observe more UCA1–hnRNP I interaction, as supported by RNA precipitation assays. Thus, this doxo-induced UCA1 is likely due to the increased stability of UCA1 once it is bound to hnRNP I.
Regulation of p27 can occur at various levels. At the transcriptional level, p27 is activated by FoxO in response to cytokines, promyelocytic leukemia proteins and nuclear Akt signaling.37 At the post-transcriptional level, p27 can be subject to proteolysis by Kip1 ubiquitination promoting complex39 or microRNA regulation.40 At the translation level, the 5′-UTR of p27 mRNA has been suggested to form a secondary structure to serve as an IRES41 and hnRNP I can interact with this IRES to promote its translation.19 In consistent with this finding, we show that UCA1 can impact this hnRNP I-mediated translation of p27, as demonstrated by luciferase reporter assays (Figure 4d). It appears that there is a competition for hnRNP I between UCA1 and p27 mRNA because ectopic expression of UCA1 causes a decrease in the p27 mRNA and hnRNP I complex (Figure 4f). Furthermore, although both UCA1 and p27 mRNA can interact with hnRNP I, they are present in separate hnRNP I complexes. Thus, it would be of interest to determine whether UCA1 and p27 interact with hnRNP I at the same binding site. Alternatively, the binding of UCA1 or p27 mRNA to hnRNP I could allosterically block subsequent binding. Therefore, our finding adds another piece (UCA1) to the p27 tumor suppressor regulatory network.
The role of hnRNP I in cancer has been well demonstrated. Of particular interest, hnRNP I has been shown, along with hnRNP A1 and hnRNP A2, to regulate the metabolic shift from oxidative phosphorylation to aerobic glycolysis by switching the splice isoforms (M1 or M2) of the glycolytic enzyme pyruvate kinase.42 It is known that normal cells express the pyruvate kinase M1 isoform (PKM1), whereas tumor cells predominantly express the M2 isoform (PKM2). The finding that overexpression of hnRNP I facilitates the production of PKM2 may underscores its oncogenic role. Furthermore, dysregulation of hnRNP I has been shown to be associated with alterations of alterative splicing and oncogenesis.43 Therefore, our study provides an additional mechanism by which hnRNP I may impact tumor cell growth.
Taken together, we demonstrate that UCA1 has an oncogenic role in breast cancer in part through the suppression of p27. We suggest that hnRNP I is an important mediator for the suppression of p27 by UCA1 (Supplementary Figure S8). Interaction of hnRNP I with UCA1 increases the stability of UCA1. Under normal situations, hnRNP I is present predominantly in the nucleus, whereas a small portion remains in the cytoplasm. However, in the presence of DNA-damaging agents such as doxo, the level of cytoplasmic hnRNP I significantly increases presumably through its phosphorylation and subcellular redistribution, as we show by hnRNP I–GFP fusion and cell fractionation experiments. This is supported by previous reports that phosphorylation impacts the subcellular localization of hnRNP I.44, 45 Protein kinase A (PKA) serves a kinase for hnRNP I phosphorylation.44 In the cytoplasm, phosphorylated hnRNP I (p-hnRNP I) is able to bind to 5′-UTR of p27 and promote its translation. At the same time, p-hnRNP I is also able to interact with UCA1. Thus, there is a competition between p27 mRNA and UCA1 for p-hnRNP I. Although this interaction can increase p27 translation, a flip side is the increased level of UCA1 at the same time. Thus, the balance between the hnRNP I–p27 mRNA complex and the hnRNP I-UCA1 is critical. Alterations in UCA1, as we detected in breast tumors, may shift this balance. When UCA1 is overexpressed, the cytoplasmic hnRNP I is recruited to UCA1 such that less hnRNP I is available to p27, leading cell cycle arrest at G1. Further characterization of these complexes will provide new insight into UCA1-mediated gene regulation in cancer and as a result, UCA1 may serve as a biomarker for breast cancer diagnosis.
Materials and Methods
Reagents
Sources of primary antibodies were: p27 and Ki-67 from Cell Signaling (Danvers, MA, USA); hnRNP I from Santa Cruz Biotechnology (Dallas, TX, USA); GAPDH from ProteinTech (Chicago, IL, USA); and β-actin from Sigma-Aldrich (St. Louis, MO, USA). Antibody against phosphorylated hnRNP I were custom made from a synthetic peptide GTKRGSDELF with Ser-16 phosphorylated (PTBP1 amino acids 11–20) by Alpha Diagnostics (San Antonio, TX, USA) using the same procedure as previously described.46 Secondary antibodies conjugated with IRDye 800CW or IRDye 680 were purchased from LI-COR Biosciences (Lincoln, NE, USA).
PCR primers were purchased from IDT (Coralville, IA, USA) (Supplementary Table S1). UCA1 siRNAs and control siRNA were purchased from ThermoFisher Scientific (Waltham, MA, USA). hnRNP I-siRNA mixture was purchased from Santa Cruz Biotechnology. Biotin-labeled UCA1-LNA probe and control oligo for in situ hybridization were purchased from Exiqon (2950 Vedbaek, Denmark). Breast cancer TMAs were purchased from US Biomax (Rockville, MD, USA).
Cell culture
Breast cancer MCF-7 and MDA-MB-231 cells (both from ATCC, Manassas, VA, USA), and HCT-116 p53-WT and HCT-116 p53-null cells, as described previously,47 were grown in RPMI 1640 from Lonza (Walkersville, MD, USA) supplemented with 10% FBS (Sigma-Aldrich), 2 mM glutamine, 100 units of penicillin/ml and 100 μg of streptomycin/ml (Lonza). Cells were incubated at 37 °C and supplemented with 5% CO2 in the humidified chamber.
LncRNA profiling
For lncRNA profiling, we used Human Disease-Related LncRNA Profiler (CAT no. RA920D, System Biosciences, Mountain View, CA, USA) consisting of 83 lncRNAs that has been described previously.48 In brief, RNA mmunoprecipitation used Magna RIP RNA-Binding Protein Immunoprecipitation Kit from Millipore (Billerica, MA, USA) and hnRNP I antibody according to the manufacturer's protocol. After the antibody was recovered by protein A+G beads (ThermoFisher Scientific, Pittsburgh, PA, USA), qRT–PCR was performed to detect lncRNA expression as described previously.48 IgG served as a control.
Transfection
Cells were transfected with siRNAs using RNAfectin reagent (Applied Biological Materials, Vancouver, Canada) or plasmid DNA using DNAfectin (Applied Biological Materials) following the manufacturer's protocol as previously described.14
Plasmid construction
PCR reactions for cloning purpose, we used Phusion enzyme (ThermoFisher Scientific, Pittsburgh, PA, USA). The entire UCA1 (1.4 kb isoform) was amplified by RT–PCR using primers UCA1-BamH I-5.1 and UCA1-EcoR I-3.1 (Supplementary Table S1), and then cloned into the expression vector pCDH-CMV-MSC-EF1-copGFP (System Biosciences) using Cold Fusion kit (System Biosciences). The same strategy was used to generate other constructs in this study, otherwise stated. To generate UCA1 RNA probes for RNA precipitation, we introduced T7 promoter sequence into the PCR products from primers. For example, to clone the full-length 1.4 kb UCA1, we used primers UCA1-T7-5.1 and UCA1-Not1-3.1. The PCR product was cloned into pCR8. The purpose of Not I site was used to linearize the plasmid before in vitro transcription. To mutagenize the potential hnRNP I-binding site, we performed two PCR reactions. The firs PCR reaction used primers UCA1-T7-5.1 and UCA1-mt-3.1A. For the second PCR reaction, we used the first PCR product as template, and primers UCA1-T7-5.1 and UCA1-mt-Not I-3.1. A dual luciferase construct carrying p27 5′-UTR was made by first PCR amplification using primers p27-5UTR-Xba I-luc-5.1, p27-5UTR-renilla-5.1, p27-5UTR-renilla-3.1 and Fluc-R-Xba I-3.1 by the same two step PCR approach, and then directly cloned into pGL3-control at Xba1 site. All PCR products were verified by DNA sequencing. Myc-tag-hnRNP I was described previously.14 GFP–hnRNP I fusion construct was provided by Dr. Xiaolong He at University of Illinois at Chicago.
Quantitative RT–PCR
LncRNA profiling by qRT–PCR was performed according to the manufacturer's protocol. Delta-delta Ct values were used to determine their relative expression as fold changes, as previously described.49
Western blot
Cells were harvested, and protein was extracted from transfected cells and quantified as previously described47 using 12% or 4∼20% polyacrylamide gradient SDS gel.
RNA precipitation
To determine whether UCA1 is associated with hnRNP I or p27 mRNA, we performed the RNA precipitation assay using biotin-labeled UCA1 RNA probe and then detected hnRNP I by western blot. In brief, the DNA fragment covering the entire 1.4 kb UCA1 sequence or deleted form was PCR-amplified using a T7-containing primers and then cloned into pCR8 (Invitrogen, Grand Island, NY, USA). In addition, another lncRNA, RoR exon 4 fragment, was also cloned as a positive control and used in precipitation experiments for comparison as described previously.14 The resultant plasmid DNA was linearized with restriction enzyme Not I which was introduced from the reverse PCR primer, and then used to synthesize RNA by T7 polymerase. The rest of the procedure was same as previously described.14
Chromogenic in situ hybridization
CISH was used to detect UCA1 in clinical specimens essentially based on a previously described method.48 A biotin-labeled antisense LNA probe was derived from nt 661∼687 of the 1.4 kb UCA1 (Supplementary Table S1). The relative signal was assessed based on the intensity as 0 (negative), same as a negative control, + (weak positive) and ++ (strong positive).
Immunohistochemistry
Immunohistochemistry (IHC) was used to detect p27 expression in breast cancer TMAs or to detect Ki-67 in xenograft tumors using the procedure as previously described.49 Relatively expression of p27 was assessed by the same procedure as for UCA1 expression.
Fluorescence in situ hybridization
Both FISH and IF stainings were used to colocalize UCA1 and hnRNPI in MCF-7 cells. FISH was performed essentially same as previously described50 except that signals were revealed by TSA Kit no. 24 with Alexa Fluor 568 (Invitrogen). Afterward, a standard IF was performed to detect phosphorylated hnRNP I as described previously.14
MTT assay
MTT assay was performed to determine the effect of UCA1 in cell growth as described previously.51 In brief, MCF-7 cells were first transfected with vector control or UCA1; control siRNA or UCA1-siRNA. The transfected cells were then seeded in 96-well plates and further cultivated for 3 more days before subjecting to MTT assays.
Luciferase assay
Luciferase assays were performed using a luciferase assay kit from Promega (Madison, WI, USA) according to the manufacturer's protocol, as previously described.49 Briefly, cells were first transfected with appropriate plasmids in 12-well plates. Then, the cells were harvested and lysed for luciferase assay 24 h after transfection. Firefly luciferase was used as a base line.
Animal work
Female nude (nu/nu) mice (4–5 weeks old) were purchased from Charles River (Wilmington, MA, USA). All animal studies were conducted in accordance with NIH animal use guidelines and a protocol approved by the UMMC Animal Care Committee. MCF-7 cells were transfected with control siRNA and UCA-siRNA, respectively. The cells at the exponential stage were harvested and were then mixed with 50% matrigel (BD Biosciences, San Jose, CA, USA). Tumor cell injection was performed as described previously.49 Paraffin-imbedded tissues were prepared for IHC staining.
Statistical analysis
Statistical analysis of data was performed using the Student's t-test. Differences with P-values less than 0.05 are considered significant.
Acknowledgments
We thank Dr. Xiaolong He at University of Illinois at Chicago for providing the GFP–hnRNP I fusion construct. This work was in part supported by NIH grant R01 CA154989 (YM).
Glossary
- CISH
chromogenic in situ hybridization
- FISH
fluorescence in situ hybridization
- IF
immunofluorescence
- hnRNP I
heterogeneous nuclear ribonucleoprotein I
- IRES
internal ribosome entry site
- lncRNA
long non-coding RNA
- PCR
polymerase chain reaction
- PRC2
polycomb repressive complex 2
- qRT–PCR
quantitative RT-PCR
- RIP
RNA immunoprecipitation
- TMA
tissue microarray
- UCA1
urothelial carcinoma-associated 1
- UTR
untranslated region
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies this paper on Cell Death and Disease website (http://www.nature.com/cddis)
Edited by G Melino
Supplementary Material
References
- Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Chinnaiyan AM. The emergence of lncRNAs in cancer biology. Cancer Discovery. 2011;1:391–407. doi: 10.1158/2159-8290.CD-11-0209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta RA, Shah N, Wang KC, Kim J, Horlings HM, Wong DJ, et al. Long non-coding RNA HOTAIR reprograms chromatin state to promote cancer metastasis. Nature. 2010;464:1071–1076. doi: 10.1038/nature08975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guttman M, Donaghey J, Carey BW, Garber M, Grenier JK, Munson G, et al. lincRNAs act in the circuitry controlling pluripotency and differentiation. Nature. 2011;477:295–300. doi: 10.1038/nature10398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khalil AM, Guttman M, Huarte M, Garber M, Raj A, Rivea Morales D, et al. Many human large intergenic noncoding RNAs associate with chromatin-modifying complexes and affect gene expression. Proc Natl Acad Sci USA. 2009;106:11667–11672. doi: 10.1073/pnas.0904715106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai MC, Manor O, Wan Y, Mosammaparast N, Wang JK, Lan F, et al. Long noncoding RNA as modular scaffold of histone modification complexes. Science. 2010;329:689–693. doi: 10.1126/science.1192002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, et al. Extensive and coordinated transcription of noncoding RNAs within cell-cycle promoters. Nat Genet. 2011;43:621–629. doi: 10.1038/ng.848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mercer TR, Dinger ME, Mattick JS. Long non-coding RNAs: insights into functions. Nat Rev Genet. 2009;10:155–159. doi: 10.1038/nrg2521. [DOI] [PubMed] [Google Scholar]
- Wang KC, Chang HY. Molecular mechanisms of long noncoding RNAs. Mol Cell. 2011;43:904–914. doi: 10.1016/j.molcel.2011.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takagi M, Absalon MJ, McLure KG, Kastan MB. Regulation of p53 translation and induction after DNA damage by ribosomal protein L26 and nucleolin. Cell. 2005;123:49–63. doi: 10.1016/j.cell.2005.07.034. [DOI] [PubMed] [Google Scholar]
- Prensner JR, Iyer MK, Balbin OA, Dhanasekaran SM, Cao Q, Brenner JC, et al. Transcriptome sequencing across a prostate cancer cohort identifies PCAT-1, an unannotated lincRNA implicated in disease progression. Nat Biotechnol. 2011;29:742–749. doi: 10.1038/nbt.1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang A, Zhou N, Huang J, Liu Q, Fukuda K, Ma D, et al. The human long non-coding RNA-RoR is a p53 repressor in response to DNA damage. Cell Res. 2013;23:340–350. doi: 10.1038/cr.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izquierdo JM, Majos N, Bonnal S, Martinez C, Castelo R, Guigo R, et al. Regulation of Fas alternative splicing by antagonistic effects of TIA-1 and PTB on exon definition. Mol Cell. 2005;19:475–484. doi: 10.1016/j.molcel.2005.06.015. [DOI] [PubMed] [Google Scholar]
- Ray PS, Grover R, Das S. Two internal ribosome entry sites mediate the translation of p53 isoforms. EMBO Rep. 2006;7:404–410. doi: 10.1038/sj.embor.7400623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang DQ, Halaby MJ, Zhang Y. The identification of an internal ribosomal entry site in the 5'-untranslated region of p53 mRNA provides a novel mechanism for the regulation of its translation following DNA damage. Oncogene. 2006;25:4613–4619. doi: 10.1038/sj.onc.1209483. [DOI] [PubMed] [Google Scholar]
- Grover R, Ray PS, Das S. Polypyrimidine tract binding protein regulates IRES-mediated translation of p53 isoforms. Cell Cycle. 2008;7:2189–2198. doi: 10.4161/cc.7.14.6271. [DOI] [PubMed] [Google Scholar]
- Cho S, Kim JH, Back SH, Jang SK. Polypyrimidine tract-binding protein enhances the internal ribosomal entry site-dependent translation of p27Kip1 mRNA and modulates transition from G1 to S phase. Mol Cell Biol. 2005;25:1283–1297. doi: 10.1128/MCB.25.4.1283-1297.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Koff A, Ohtsuki M, Polyak K, Roberts JM, Massague J. Negative regulation of G1 in mammalian cells: inhibition of cyclin E-dependent kinase by TGF-beta. Science. 1993;260:536–539. doi: 10.1126/science.8475385. [DOI] [PubMed] [Google Scholar]
- Catzavelos C, Bhattacharya N, Ung YC, Wilson JA, Roncari L, Sandhu C, et al. Decreased levels of the cell-cycle inhibitor p27Kip1 protein: prognostic implications in primary breast cancer. Nat Med. 1997;3:227–230. doi: 10.1038/nm0297-227. [DOI] [PubMed] [Google Scholar]
- Han S, Park K, Kim HY, Lee MS, Kim HJ, Kim YD. Reduced expression of p27Kip1 protein is associated with poor clinical outcome of breast cancer patients treated with systemic chemotherapy and is linked to cell proliferation and differentiation. Breast Cancer Res Treatment. 1999;55:161–167. doi: 10.1023/a:1006258222233. [DOI] [PubMed] [Google Scholar]
- Hengst L, Reed SI. Translational control of p27Kip1 accumulation during the cell cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
- Wander SA, Zhao D, Slingerland JM. p27: a barometer of signaling deregulation and potential predictor of response to targeted therapies. Clin Cancer Res. 2011;17:12–18. doi: 10.1158/1078-0432.CCR-10-0752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang XS, Zhang Z, Wang HC, Cai JL, Xu QW, Li MQ, et al. Rapid identification of UCA1 as a very sensitive and specific unique marker for human bladder carcinoma. Clinl Cancer Res. 2006;12:4851–4858. doi: 10.1158/1078-0432.CCR-06-0134. [DOI] [PubMed] [Google Scholar]
- Tsang WP, Wong TW, Cheung AH, Co CN, Kwok TT. Induction of drug resistance and transformation in human cancer cells by the noncoding RNA CUDR. RNA. 2007;13:890–898. doi: 10.1261/rna.359007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng S, Gray EE, Chawla G, Porse BT, O'Dell TJ, Black DL.PSD-95 is post-transcriptionally repressed during early neural development by PTBP1 and PTBP2 Nat Neurosci 201215381–388.S381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheung HC, Hai T, Zhu W, Baggerly KA, Tsavachidis S, Krahe R, et al. Splicing factors PTBP1 and PTBP2 promote proliferation and migration of glioma cell lines. Brain. 2009;132 (Pt 8:2277–2288. doi: 10.1093/brain/awp153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cobbold LC, Wilson LA, Sawicka K, King HA, Kondrashov AV, Spriggs KA, et al. Upregulated c-myc expression in multiple myeloma by internal ribosome entry results from increased interactions with and expression of PTB-1 and YB-1. Oncogene. 2010;29:2884–2891. doi: 10.1038/onc.2010.31. [DOI] [PubMed] [Google Scholar]
- Xue Y, Ouyang K, Huang J, Zhou Y, Ouyang H, Li H, et al. Direct conversion of fibroblasts to neurons by reprogramming PTB-regulated microRNA circuits. Cell. 2013;152:82–96. doi: 10.1016/j.cell.2012.11.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amaral PP, Clark MB, Gascoigne DK, Dinger ME, Mattick JS. lncRNAdb: a reference database for long noncoding RNAs. Nucleic Acids Res. 2011;39 (Database issue:D146–D151. doi: 10.1093/nar/gkq1138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tillmar L, Carlsson C, Welsh N. Control of insulin mRNA stability in rat pancreatic islets. Regulatory role of a 3'-untranslated region pyrimidine-rich sequence. J Biol Chem. 2002;277:1099–1106. doi: 10.1074/jbc.M108340200. [DOI] [PubMed] [Google Scholar]
- Wang MJ, Lin S. A region within the 5'-untranslated region of hypoxia-inducible factor-1alpha mRNA mediates its turnover in lung adenocarcinoma cells. J Biol Chem. 2009;284:36500–36510. doi: 10.1074/jbc.M109.008904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartek J, Iggo R, Gannon J, Lane DP. Genetic and immunochemical analysis of mutant p53 in human breast cancer cell lines. Oncogene. 1990;5:893–899. [PubMed] [Google Scholar]
- Polyak K, Lee MH, Erdjument-Bromage H, Koff A, Roberts JM, Tempst P, et al. Cloning of p27Kip1, a cyclin-dependent kinase inhibitor and a potential mediator of extracellular antimitogenic signals. Cell. 1994;78:59–66. doi: 10.1016/0092-8674(94)90572-x. [DOI] [PubMed] [Google Scholar]
- Chu I, Sun J, Arnaout A, Kahn H, Hanna W, Narod S, et al. p27 phosphorylation by Src regulates inhibition of cyclin E-Cdk2. Cell. 2007;128:281–294. doi: 10.1016/j.cell.2006.11.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chu IM, Hengst L, Slingerland JM. The Cdk inhibitor p27 in human cancer: prognostic potential and relevance to anticancer therapy. Nat Rev Cancer. 2008;8:253–267. doi: 10.1038/nrc2347. [DOI] [PubMed] [Google Scholar]
- Oberstrass FC, Auweter SD, Erat M, Hargous Y, Henning A, Wenter P, et al. Structure of PTB bound to RNA: specific binding and implications for splicing regulation. Science. 2005;309:2054–2057. doi: 10.1126/science.1114066. [DOI] [PubMed] [Google Scholar]
- Kotoshiba S, Kamura T, Hara T, Ishida N, Nakayama KI. Molecular dissection of the interaction between p27 and Kip1 ubiquitylation-promoting complex, the ubiquitin ligase that regulates proteolysis of p27 in G1 phase. J Biol Chem. 2005;280:17694–17700. doi: 10.1074/jbc.M500866200. [DOI] [PubMed] [Google Scholar]
- le Sage C, Nagel R, Agami R. Diverse ways to control p27Kip1 function: miRNAs come into play. Cell Cycle. 2007;6:2742–2749. doi: 10.4161/cc.6.22.4900. [DOI] [PubMed] [Google Scholar]
- Gopfert U, Kullmann M, Hengst L. Cell cycle-dependent translation of p27 involves a responsive element in its 5'-UTR that overlaps with a uORF. Hum Mol Genet. 2003;12:1767–1779. doi: 10.1093/hmg/ddg177. [DOI] [PubMed] [Google Scholar]
- David CJ, Chen M, Assanah M, Canoll P, Manley JL. HnRNP proteins controlled by c-Myc deregulate pyruvate kinase mRNA splicing in cancer. Nature. 2010;463:364–368. doi: 10.1038/nature08697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He X, Pool M, Darcy KM, Lim SB, Auersperg N, Coon JS, et al. Knockdown of polypyrimidine tract-binding protein suppresses ovarian tumor cell growth and invasiveness in vitro. Oncogene. 2007;26:4961–4968. doi: 10.1038/sj.onc.1210307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc Natl Acad Sci USA. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knoch KP, Meisterfeld R, Kersting S, Bergert H, Altkruger A, Wegbrod C, et al. cAMP-dependent phosphorylation of PTB1 promotes the expression of insulin secretory granule proteins in beta cells. Cell Metab. 2006;3:123–134. doi: 10.1016/j.cmet.2005.12.008. [DOI] [PubMed] [Google Scholar]
- Xie J, Lee JA, Kress TL, Mowry KL, Black DL. Protein kinase A phosphorylation modulates transport of the polypyrimidine tract-binding protein. Proc Natl Acad Sci USA. 2003;100:8776–8781. doi: 10.1073/pnas.1432696100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sachdeva M, Zhu S, Wu F, Wu H, Walia V, Kumar S, et al. p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc Natl Acad Sci USA. 2009;106:3207–3212. doi: 10.1073/pnas.0808042106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Q, Huang J, Zhou N, Zhang Z, Zhang A, Lu Z, et al. LncRNA loc285194 is a p53-regulated tumor suppressor. Nucleic Acids Res. 2013;41:4976–4987. doi: 10.1093/nar/gkt182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Si ML, Zhu S, Wu H, Lu Z, Wu F, Mo YY. miR-21-mediated tumor growth. Oncogene. 2007;26:2799–2803. doi: 10.1038/sj.onc.1210083. [DOI] [PubMed] [Google Scholar]
- Gupta A, Mo YY. Detection of microRNAs in cultured cells and paraffin-embedded tissue specimens by in situ hybridization. Methods Mol Biol. 2011;676:73–83. doi: 10.1007/978-1-60761-863-8_6. [DOI] [PubMed] [Google Scholar]
- Wu F, Chiocca S, Beck WT, Mo YY. Gam1-associated alterations of drug responsiveness through activation of apoptosis. Mol Cancer Ther. 2007;6:1823–1830. doi: 10.1158/1535-7163.MCT-06-0771. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.