Fig. 1.
The rankprop algorithm. Given a set of objects (in this case, proteins) X = x1,..., xm, let x1 be the query and x2,..., xm be the database (targets) we would like to rank. Let K be the matrix of object–object similarities, i.e., Kij gives a similarity score between xi and xj, with K normalized so that 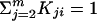 for all i. For computational efficiency, we set K1i = Ki1 for all i, so that we can compute weights involving the query using a single execution of psi-blast. Let yi, i = 2,..., m, be the initial ranking “score” of a target. In practice, for efficiency, the algorithm is terminated after a fixed number I of iterations, and yi(I) is used as an approximation of
for all i. For computational efficiency, we set K1i = Ki1 for all i, so that we can compute weights involving the query using a single execution of psi-blast. Let yi, i = 2,..., m, be the initial ranking “score” of a target. In practice, for efficiency, the algorithm is terminated after a fixed number I of iterations, and yi(I) is used as an approximation of 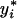 . The parameter α ∈ [0,1] is set a priori by the user. For α = 0, no global structure is found, and the algorithm's output is just the ranking according to the original distance metric. These experiments use α = 0.95, looking for clear cluster structure in the data.
. The parameter α ∈ [0,1] is set a priori by the user. For α = 0, no global structure is found, and the algorithm's output is just the ranking according to the original distance metric. These experiments use α = 0.95, looking for clear cluster structure in the data.

