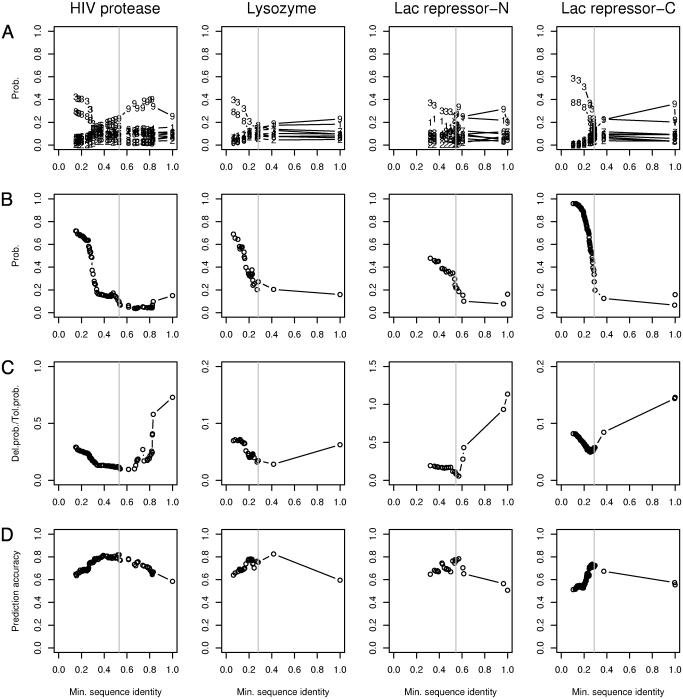
Fig. 1.
Shown are inferences of which protein sequences in a multiple sequence alignment are functionally related to a reference sequence. The plots correspond to the reference protein domains HIV protease (residues 4–99), T4 lysozyme (residues 24–148), Lac-N (residues 2–29), and Lac-C (residues 68–327). The abscissa of each plot is the minimal fraction of amino acids shared with the reference sequence for each subalignment of the full multiple sequence alignment extracted from the Pfam database. (A) The contribution from each of the nine Dirichlet components from the Blocks9 mixture. The numerical plot symbols refer to the contributing components. (B) The sum of the contributions from components 3 and 8 from A. (C) The ratio of posterior estimates of amino acids experimentally determined to be either functionally deleterious or functionally tolerated. The minimum of each curve suggests which subalignments optimally inform the functional predictions in D. (D) Overall prediction accuracy of tolerated vs. deleterious amino acid substitutions. The gray vertical line in each plot indicates the subalignment selected for representing each query sequence. The subalignment sequence identity cutoffs are 0.531 for HIV protease (although a larger rise occurs at ≈0.3, we interpret the smaller rise at ≈0.5 as indicating some level of functional divergence from the query sequence), 0.281 for T4 lysozyme, 0.545 for Lac-N, and 0.289 for Lac-C.