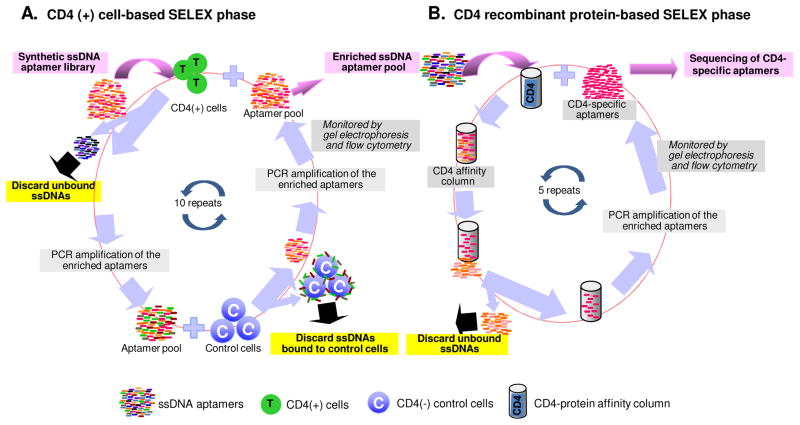
Fig. 1. Hybrid SELEX approaches to develop CD4-specific aptamers.
The hybrid SELEX protocol included a two-step enrichment process that combines CD4-positive T-cell-based enrichment with biomarker-based selection. (A) CD4-binding aptamers were selected from a synthetic single strand DNA library with CD4-positive T cells. After PCR amplification of the enriched aptamers, negative selection, using CD4-negative control cells, was carried out and aptamers that non-specifically bind to cellular components were discarded. Seven rounds of positive and three negative selections with PCR amplification were performed. (B) Aptamers able to bind CD4-postive cells were then subjected to further enrichment with protein-based selection. Aptamers were added to a CD4 affinity column and unbound ssDNAs are discarded. PCR amplification of the bound aptamers was then performed. Protein-based selection was repeated 5 times. The final aptamer pool was amplified, sequenced and consists of CD4-specific aptamers.