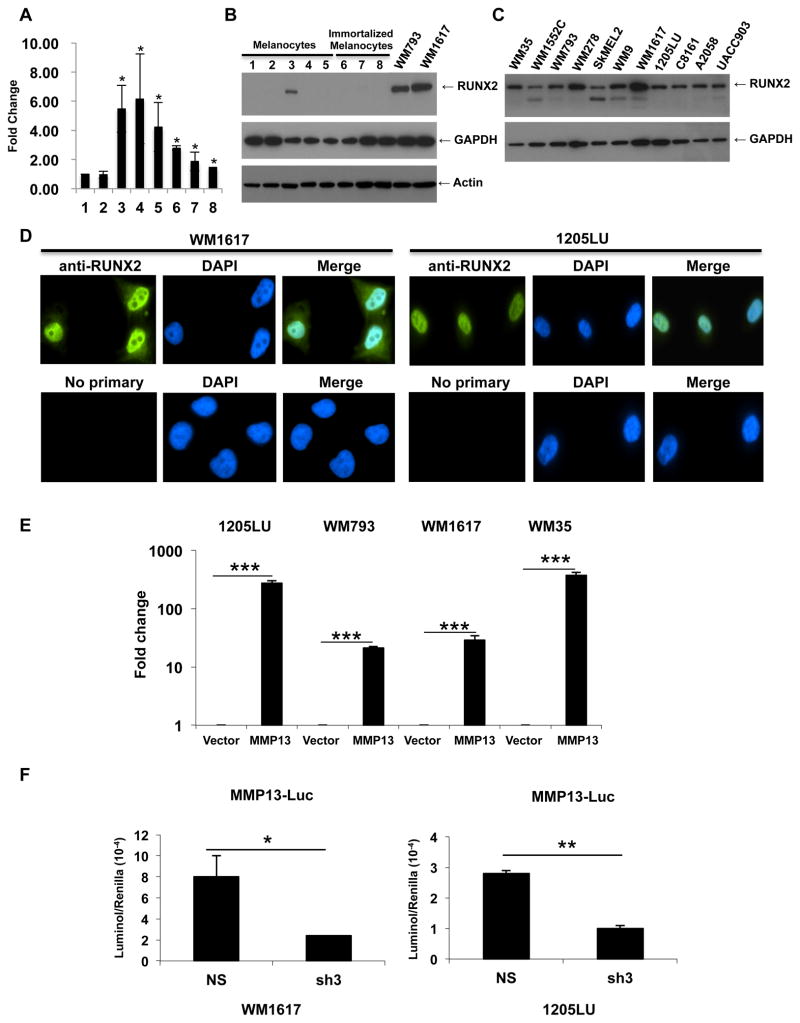
Figure 1. Transcriptionally active RUNX2 is overexpressed in melanoma cell lines as compared with melanocytes.
A: Quantitative PCR analysis of RUNX2 expression in independent primary cultures of melanocytes (1,2) and six melanoma cell lines 3: WM793; 4: C8161; 5: 1205LU; 6: WM9; 7: UACC903; 8: WM1617. *: P < 0.05 as compared with melanocytes (1). B: Immunoblot analysis of RUNX2 expression in the primary cultures of melanocytes AG22151: 1 and 2 (two different lysate preparations), AG22173: 3 and 4 (two different lysate preparations) and FOM-136-1: 5; in immortalized melanocytes: 6–8 (three different lysate preparations); and in the human melanoma cell lines WM793 and WM1617. GAPDH and Actin were used for normalization. C. Immunoblot analysis of RUNX2 in human melanoma cell lines. GAPDH was used for normalization. D. Immunofluorescence analysis of RUNX2 in the two human melanoma cell lines WM1617 and 1205LU. E. 1205LU, WM793, WM1617 and WM35 melanoma cells were transfected with the empty PGL3 vector (Vector) or the vector carrying the MMP13 promoter fused to the luciferase gene (MMP13), along with the Renilla plasmid. 24 hours post-transfection, firefly luciferase and renilla activities were measured and relative light units (RLU) were normalized to the renilla units per sample. The values for luciferase activity in the presence of MMP13-luciferase vector are reported as relative fold change compared to the transfection with vector control (set to 1). This figure is representative of three independent experiments. *** indicate p < 0.001 compared with the vector control. F. WM1617 (Left) and 1205LU (Right) melanoma cells were co-transfected with non-silencing ShRNA (NS) or shRUNX2-3 (Sh3) lentiviral plasmids along with the PGL3-MMP13-luc and renilla (phRL-CMV) plasmids. Firefly luciferase and renilla activities were measured 24 hours post-transfection, and relative light units (RLU) were normalized to the renilla units per sample. This figure is representative of three independent experiments with p values indicated. * indicates p < 0.05 and ** indicate p < 0.01 compared with the non-silencing (NS) vector.