Figure 3.
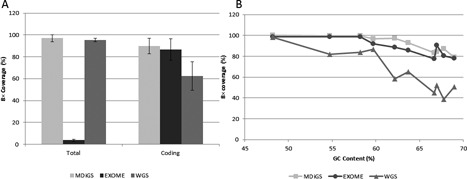
MDiGS coverage of target regions. (A) Total coverage of BAC-targeted coding and non-coding regions were equivalent to whole genome sequencing and coverage of coding regions were equivalent to exome-captured sequencing and superior to whole genome sequencing. (B) Overall coverage of GC-rich regions decreased as GC content increases in all three platforms (MDiGS, exome sequencing and whole genome sequencing). Both BAC- and exome-capture-based methods performed better than whole genome sequencing for high GC-rich genes.
