Figure 1.
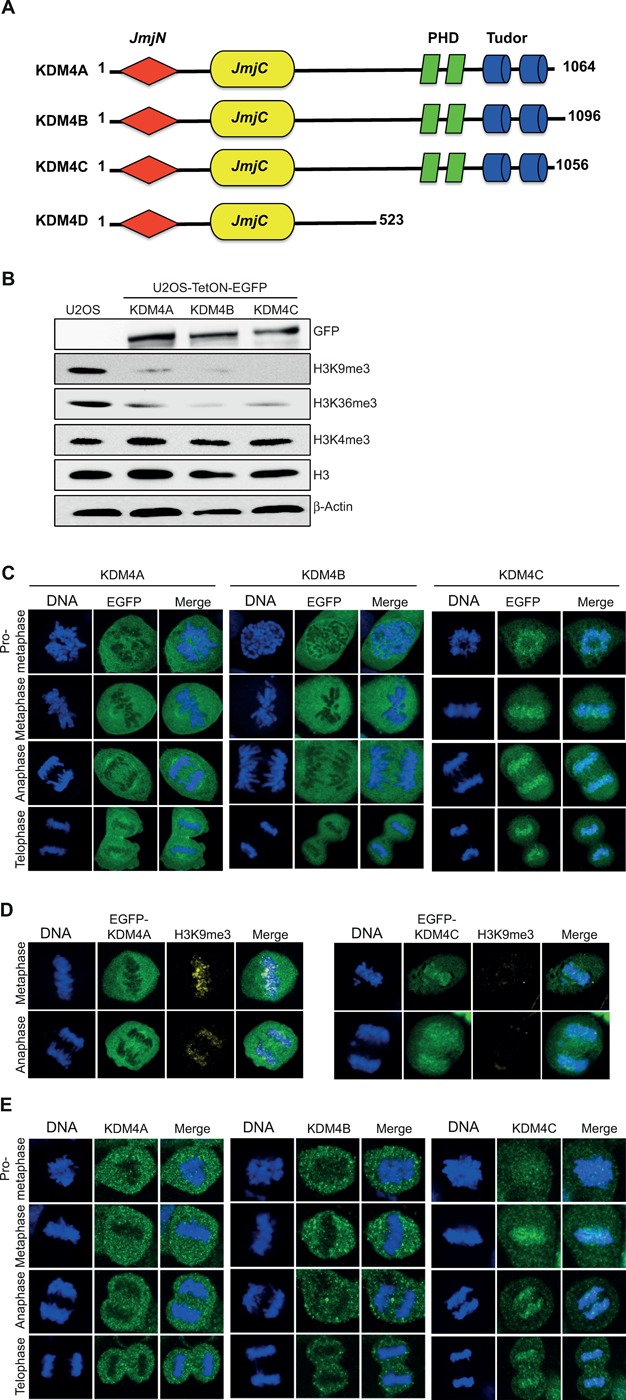
Differential localization of EGFP-KDM4A-C fusions during mitosis. (A) Schematic structure representation of the KDM4A-D histone demethylases. The KDM4 family consists of four members: KDM4A, KDM4B, KDM4C and KDM4D. All members, except KDM4D, share, in addition to JmjC and JmjN, two PHD and two Tudor domains. KDM4A-C schematic was built using the SMART software (http://smart.embl-heidelberg.de). (B) Western blot analysis shows that U2OS-TetON cells express comparable levels of KDM4A-C proteins that demethylate H3K9me3 and H3K36me3 but not H3K4me3. Cells were treated with Dox for 36 h and protein lysates were prepared using hot-lysis and immunoblotted with the indicated antibodies. (C) Representative images showing the localization of EGFP-KDM4A-C fusions from prometaphase to telophase. U2OS-Tet-ON-EGFP-KDM4A-C cells were treated with Dox to induce the expression of EGFP-KDM4A-C fusions (green). Cells were stained with DAPI (blue). KDM4A and KDM4B are excluded from chromatin while KDM4C is associated with mitotic chromatin. Twenty to thirty cells were counted for each of the mitotic stages expressing either KDM4A or KDM4B. 0ver 100 cells were counted for mitotic cells expressing EGFP-KDM4C. (D) Overexpression of EGFP-KDM4C, but not EGFP-KDM4A, leads to a severe reduction in H3K9me3 levels. U2OS-Tet-ON-EGFP-KDM4A and U2OS-Tet-ON-EGFP-KDM4C cells were treated with Dox for 36 h, fixed and subjected to immunofluorescence analysis using H3K9me3 antibody (yellow). DNA is stained with DAPI (blue) and EGFP-KDM4A and EGFP-KDM4C are in green. Results shown in (C) and (D) are typical of at least two independent experiments. (E) Representative images showing the localization of the endogenous KDM4A-C proteins from prometaphase to telophase. MCF7 cells were fixed and subjected to immunofluorescence analysis using KDM4A-C antibodies.
