Figure 6.
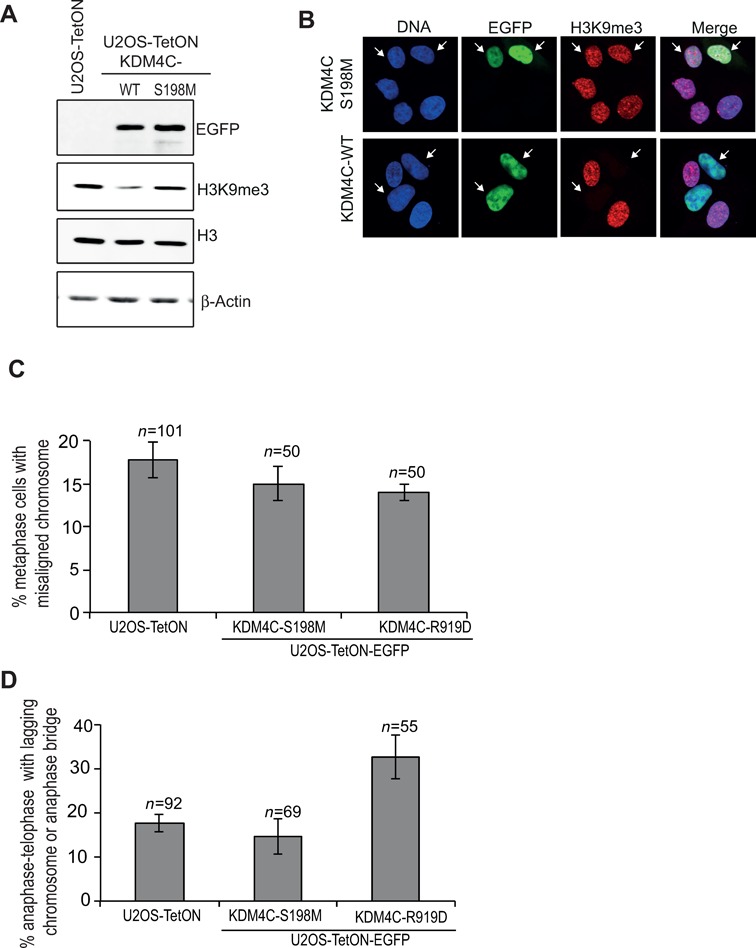
KDM4C demethylase activity and mitotic localization affect the fidelity of chromosome segregation during mitosis. (A) and (B) show the effect of S198M mutation on the demethylase activity of KDM4C protein. (A) western blot analysis showing that overexpression of EGFP-KDM4C-S198M has no detectable effect on the levels of H3K9me3. Protein extracts were prepared from U2OS-TetON cells expressing either EGFP-KDM4C-WT or EGFP-KDM4C-S198M and immunoblotted using the indicated antibodies. (B) Immunofluorescence analysis of U2OS-TetON expressing either EGFP-KDM4C-WT (bottom) or EGFP-KDM4C-S198M (top). Cells were stained for H3K9me3 (red). DNA is stained with DAPI (blue), and the EGFP-KDM4C is in green. (C) and (D) Histograms showing the percentage of metaphases with misaligned chromosomes (B) and anaphase–telophase cells that exhibit lagging chromosomes or anaphase–telophase bridges (C). U2OS-TetON cells expressing EGFP-KDM4C-S198M or EGFP-KDM4C-R919D were subjected to immunofluorescence and mitotic cells were acquired using confocal microscope. n, number of mitotic cells counted. Error bars represent standard deviation from two independent experiments.
