Figure 3.
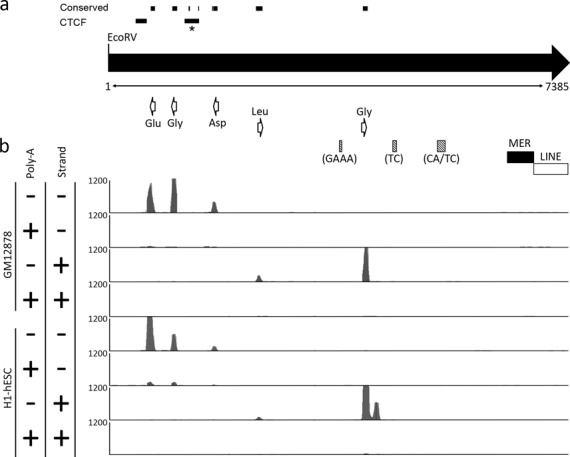
Characterization of a single repeat unit from the human chromosome 1q23.3 tRNA tandem repeat: genomic features and transcriptional units. (a) Schematic map of a single repeat unit, represented by the right-facing black arrow as defined by the periodicity of the restriction endonuclease EcoRV. The size of the repeat is indicated in bp immediately below the black arrow. The location and direction of transcription of tRNA genes are indicated by the open white arrows. The location of microsatellite repeats are indicated by the shaded gray boxes and the sequence composition of the repeat units indicated below each in brackets. The black and white boxes indicate the location of a MER and LINE element, respectively. Regions of conserved DNA sequence with the mouse repeat are indicated above the monomer, as are the location of two CTCF peaks. The peak marked with the * corresponds to the CTCF motif that is shared with mouse. (b) Representation of transcripts originating from a single repeat unit using data obtained from the ENCODE project (34). Image is adapted from the UCSC Genome Browser (www.genome.ucsc.edu) (26); build GRCh37/hg19 showing the track for Long RNA-seq from ENCODE/Cold Spring Harbor Lab. Data from the LCL GM12878 and hESC cell line H1 are indicated to the left. Polyadenylated RNA and nonpolyadenylated RNA are indicated by the ‘+’ and ‘−’ symbols, as are transcripts originating from the sense (+) and antisense (−) strands. Each track shows a vertical viewing range of 1200 reads.
