Figure 8.
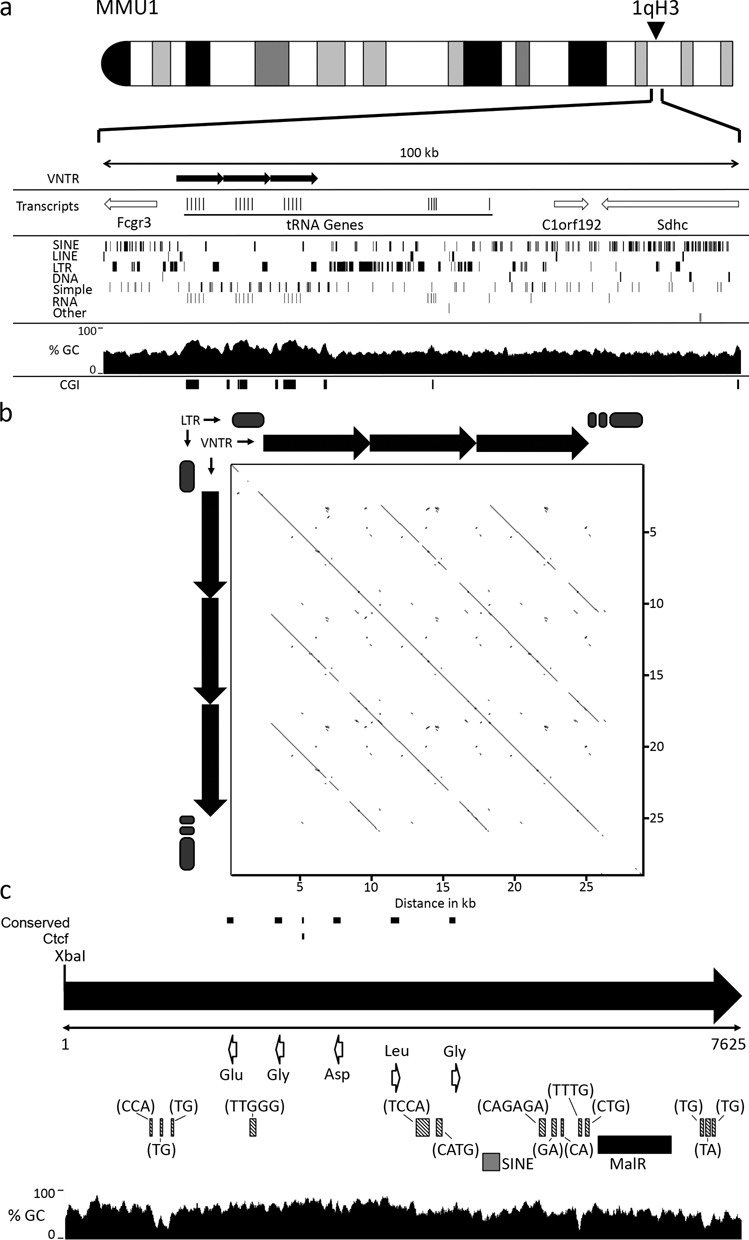
Characterization of the mouse tRNA cluster. (a) Ideogram of mouse chromosome 1, indicating the approximate location of the tRNA tandem repeat at 1qH3. Immediately below is schematic map showing a 100-kb genomic window in the vicinity of the tRNA cluster (corresponding to 172,981,160–173,081,065 of mouse chromosome 1, GRCm38/mm10). The location of the VNTR is indicated in the top section by the black right-facing arrows. Transcripts from the interval are indicated in the second section. Open arrows indicate the genomic coverage of the transcript and direction of transcription. Beneath this is a map showing the location of the indicated repeat types (left side labels). The next section shows a plot of GC percentage across the interval. The final section shows the location of CGI indicated by solid black boxes. (b) Pair-wise alignment of the VNTR and flanking LTR elements using YASS (www.http://bioinfo.lifl.fr/yass/index.php) corresponding to 172,990,701–173,020,200 of mouse chromosome 1 (GRCm38/mm10). The location of individual VNTR repeat units are represented above and to the left of the plot by black arrows. Gray oval blocks represent the location of LTRs that are excluded from the plot, indicated by gaps in the diagonal line. (c) Schematic map of a single repeat unit, represented by the right-facing black arrow as defined by the periodicity of the restriction endonuclease XbaI, corresponding to 173,000,076–173,007,700 of mouse chromosome 1 (GRC38/mm10). The size of the repeat is indicated in bp immediately below the black arrow. DNA sequences that are conserved with the human repeat are indicated as black lines above the black arrow, as is the conserved Ctcf binding motif. The location and direction of transcription of tRNA genes are indicated by the open white arrows. The location of microsatellite repeats are indicated by the shaded gray boxes and the sequence composition of the repeat units indicated in brackets. The black and gray boxes indicate the location of a MalR and SINE element, respectively.
