Figure 3.
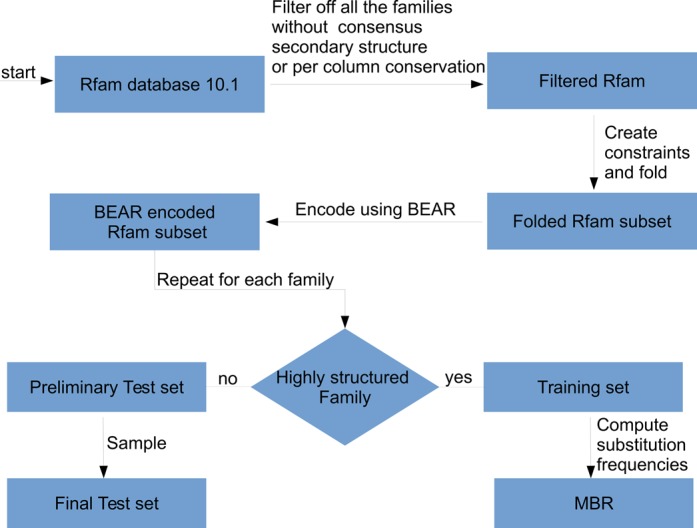
MBR construction and testing pipeline. We started from Rfam 10.1, selecting families for which a consensus structure and a per-column conservation is reported. Conserved positions in the Rfam multiple alignments were used to select structural constraints that guided RNAfold. The resulting secondary structures for each member of the Rfam families were converted into BEAR. A set of Rfam families having high density of SSEs was used to compute SSEs substitution frequencies and build the MBR (the training set). From the remaining families, pairwise alignments were randomly sampled and used to test the ability of MBR in reconstructing the alignment.
