Figure 6.
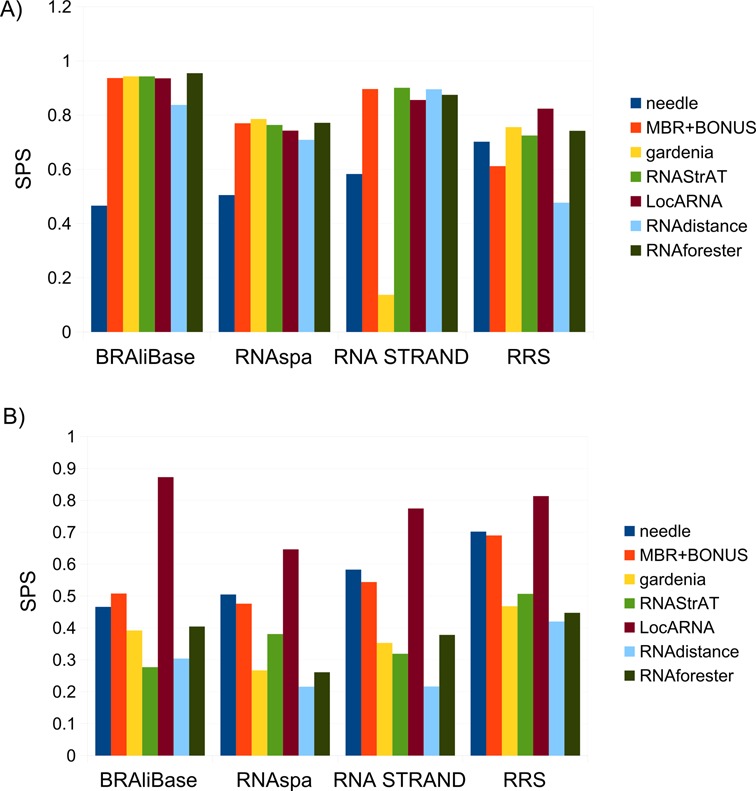
(A) Alignment performance of the seven tested methods on the four different data sets. We evaluated seven different alignment methods, from left to right: a sequence-based Needleman–Wunsch (needle), our modified Needleman–Wunsch using the MBR including the sequence BONUS (MBR+BONUS), then gardenia, RNAStrAT, LocARNA, RNAdistance and RNAforester. All methods were tested on the four employed data sets; (B) Alignment performance of the five tested methods when secondary structures are predicted using RNAfold.
