Figure 5.
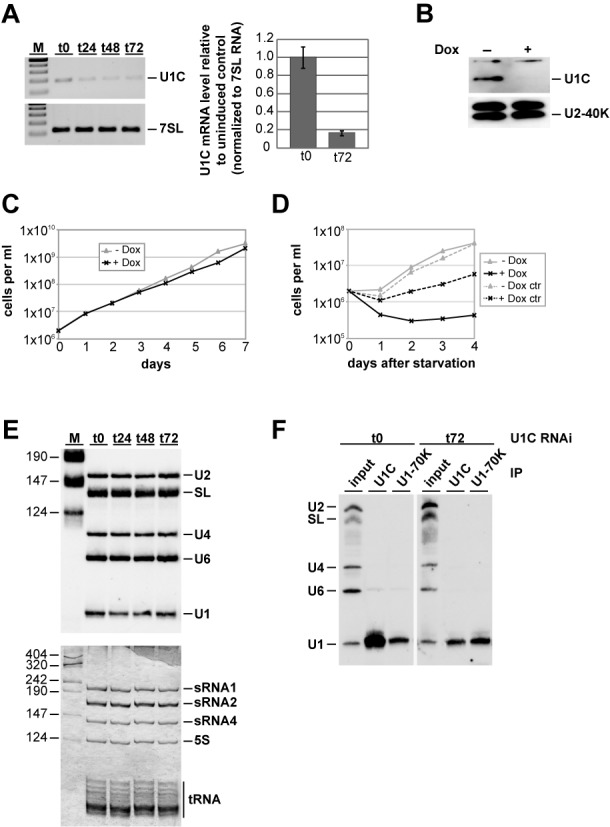
Effects of RNAi-mediated depletion of Trypanosoma brucei U1C on growth and U1 snRNP integrity. (A, B) RNAi-mediated knockdown of U1C expression. 24, 48 or 72 h after RNAi induction (as indicated), RNA was analyzed by semiquantitative RT-PCR (top) and real-time PCR (bottom). In addition, RNA from uninduced cells (t0) was included, and, as a control, 7SL RNA was measured from the same RNA samples (A). M, markers (100, 200, 300, 400 and 500 bp). In parallel, U1C protein was detected by immunoblotting with affinity-purified polyclonal antibodies, using U2-40K protein as a control (B). (C) Growth curve of a representative clonal procyclic T. brucei cell line, in which RNAi against the U1C mRNA was induced. Cells were grown for 7 days in the absence (−Dox; gray line with triangles) or in the presence of doxycyline (+Dox; black line with crosses), counted every day, and diluted back to 2 × 106 cells/ml. (D) Recovery of procyclic cells with or without induction of U1C RNAi from starvation stress. Cells were grown for 1 day in the absence (−Dox) or in the presence of 1 mg/ml doxycycline (+Dox). As a control for growth effects of doxycycline, control cells (ctr) were grown for 1 day under the same conditions. 2 × 106 cells/ml were collected, washed twice in phosphate-buffered saline (PBS), resuspended in the original volume of PBS, and incubated at 27°C for 2 h. After starvation stress, cells were harvested again, resuspended in the original volume SDM-79, and grown in the absence (−Dox; gray line with triangles) or in the presence of doxycyline (+Dox; black line with crosses). (E) U1C knockdown does not affect snRNA steady-state levels. From uninduced cells (t0) and 24, 48 and 72 h after silencing of U1C expression, equal amounts of RNA were analyzed by northern blot hybridization, using a mixed probe for U2, SL, U4, U6 and U1 snRNAs (top panel). As a loading control, equal amounts of RNAs were detected by silver staining (bottom panel). Positions of snRNAs, ribosomal RNAs and tRNAs are marked on the right. M, markers (in nucleotides). (F) Immunoprecipitation analysis of U1 snRNPs upon U1C knockdown. From uninduced cells (t0) and 72 h after U1C knockdown (t72), whole-cell extracts were prepared and subjected to immunoprecipitation (IP), using anti-U1C or U1-70K antibodies (as indicated). Copurifying RNAs were analyzed by northern blotting, using a mixed probe for U2, SL, U4, U6 and U1 snRNAs (positions indicated on the left). For comparison, 5% of each input is shown (lanes ‘input’).
