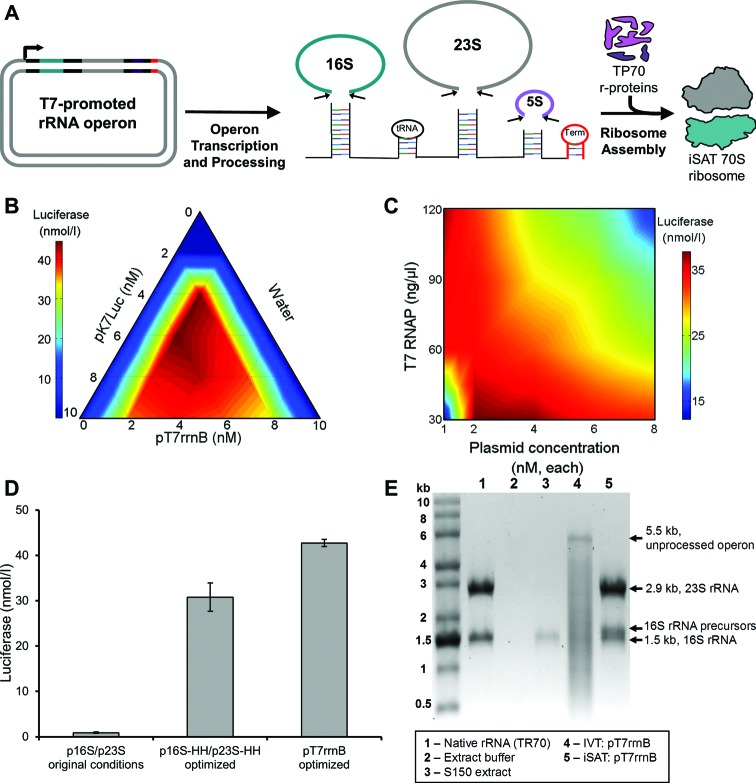
Figure 4.
Component optimization for iSAT system utilizing T7-promoted rrnB operon. (A) Diagram of expected transcription and processing of rrnB operon encoding 16S, 23S and 5S rRNA from a T7-promoted plasmid. Expected processing of transcribed RNA includes cleavage of rRNA by RNases present in S150 extract to allow for 70S ribosome assembly. All steps occur in isothermal iSAT reactions (37°C), and resulting ribosomes are assessed for activity by measuring reporter protein production, as depicted in Figure 1. (B) Ratio optimization of co-transcribed plasmids included in operon-based iSAT reactions for luciferase production. (C) Ratio optimization of T7 RNAP and plasmid concentration in operon-based iSAT reactions for luciferase production. T7 RNAP was diluted with storage buffer to maintain equivalent salt and buffer concentrations in all reactions. The plasmids pK7Luc and pT7rrnB were mixed at an equimolar ratio as determined from (B). For panels (B) and (C), values show average luciferase concentrations above background for at least two independent reactions. (D) Comparison of luciferase production for iSAT reactions utilizing p16S/p23S, p16S-HH/p23S-HH or pT7rrnB. Values show average luciferase concentrations above background with error bars representing s.d. for at least three independent reactions. (E) RNA denaturing gel of iSAT reactions utilizing pT7rrnB. The in vitro transcribed (IVT) reaction (Lane 4) included all components of iSAT reactions except S150 extract, which was replaced with extract buffer to maintain salt concentrations. TR70 control lane includes 1 μg RNA.